Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
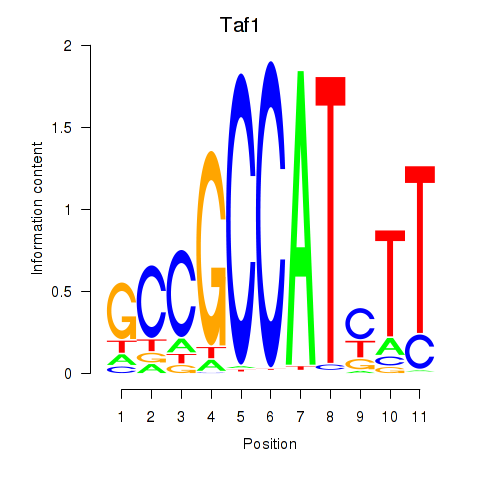
Results for Taf1
Z-value: 3.63
Transcription factors associated with Taf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Taf1
|
ENSMUSG00000031314.11 | TATA-box binding protein associated factor 1 |
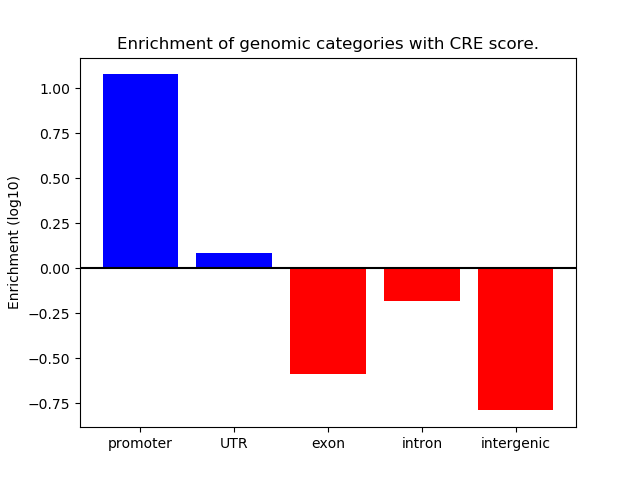
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_101532549_101532710 | Taf1 | 105 | 0.960522 | 0.94 | 5.1e-03 | Click! |
| chrX_101532903_101533136 | Taf1 | 260 | 0.901826 | -0.17 | 7.5e-01 | Click! |
| chrX_101603820_101603987 | Taf1 | 13356 | 0.160129 | 0.12 | 8.3e-01 | Click! |
Activity of the Taf1 motif across conditions
Conditions sorted by the z-value of the Taf1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
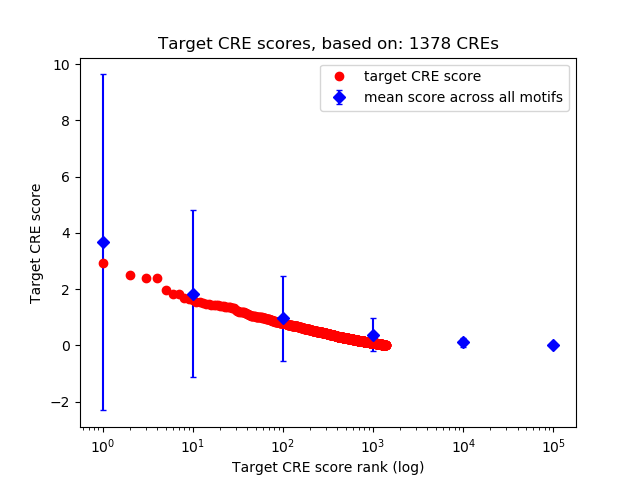
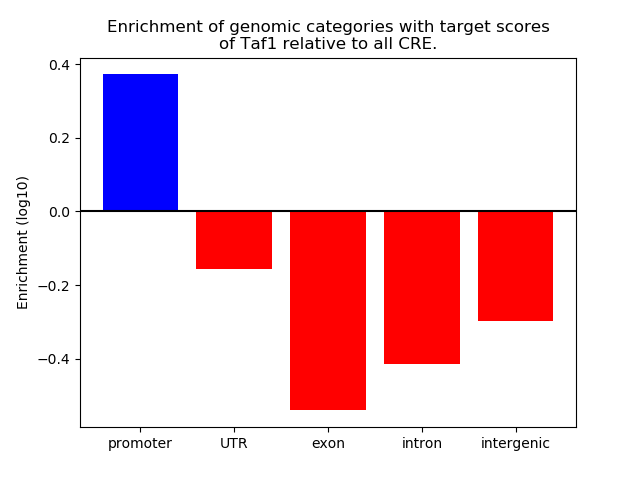
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_35022239_35022407 | 2.92 |
Hacd2 |
3-hydroxyacyl-CoA dehydratase 2 |
105 |
0.89 |
| chr1_171222423_171222787 | 2.49 |
Tomm40l |
translocase of outer mitochondrial membrane 40-like |
91 |
0.91 |
| chr16_93832267_93832500 | 2.41 |
Morc3 |
microrchidia 3 |
116 |
0.88 |
| chr7_6727806_6728072 | 2.38 |
Peg3 |
paternally expressed 3 |
2480 |
0.14 |
| chrX_94122302_94122676 | 1.96 |
Zfx |
zinc finger protein X-linked |
411 |
0.86 |
| chr16_33062416_33062582 | 1.82 |
Lmln |
leishmanolysin-like (metallopeptidase M8 family) |
22 |
0.96 |
| chr7_28338746_28338909 | 1.81 |
Supt5 |
suppressor of Ty 5, DSIF elongation factor subunit |
81 |
0.92 |
| chr2_164879263_164879427 | 1.69 |
Pcif1 |
PDX1 C-terminal inhibiting factor 1 |
12 |
0.95 |
| chr1_157080183_157080500 | 1.66 |
Gm28694 |
predicted gene 28694 |
26307 |
0.14 |
| chr14_55015448_55015599 | 1.61 |
Ngdn |
neuroguidin, EIF4E binding protein |
69 |
0.92 |
| chr17_74078571_74079138 | 1.56 |
Gm49880 |
predicted gene, 49880 |
192 |
0.48 |
| chr7_99980419_99980597 | 1.55 |
Rnf169 |
ring finger protein 169 |
60 |
0.96 |
| chr11_107132103_107132265 | 1.49 |
Bptf |
bromodomain PHD finger transcription factor |
57 |
0.97 |
| chr3_95624833_95625010 | 1.47 |
Ensa |
endosulfine alpha |
72 |
0.94 |
| chr8_69998729_69998914 | 1.47 |
Gatad2a |
GATA zinc finger domain containing 2A |
2437 |
0.18 |
| chr12_100187457_100187764 | 1.44 |
Gm10433 |
predicted gene 10433 |
358 |
0.62 |
| chr1_133302040_133302233 | 1.44 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
4185 |
0.14 |
| chr1_182758644_182758936 | 1.42 |
Susd4 |
sushi domain containing 4 |
5070 |
0.21 |
| chr5_35719956_35720115 | 1.42 |
Sh3tc1 |
SH3 domain and tetratricopeptide repeats 1 |
797 |
0.57 |
| chr15_6562479_6562656 | 1.40 |
Fyb |
FYN binding protein |
6960 |
0.23 |
| chrX_151803526_151803760 | 1.39 |
Huwe1 |
HECT, UBA and WWE domain containing 1 |
15 |
0.97 |
| chrX_50620045_50620213 | 1.38 |
Firre |
functional intergenic repeating RNA element |
14937 |
0.26 |
| chr2_94406648_94406807 | 1.37 |
Ttc17 |
tetratricopeptide repeat domain 17 |
38 |
0.91 |
| chr11_104442214_104442368 | 1.37 |
Kansl1 |
KAT8 regulatory NSL complex subunit 1 |
0 |
0.97 |
| chr1_171329168_171329338 | 1.37 |
Dedd |
death effector domain-containing |
108 |
0.89 |
| chr17_35164544_35164698 | 1.36 |
Prrc2a |
proline-rich coiled-coil 2A |
214 |
0.62 |
| chr8_20297375_20297660 | 1.34 |
6820431F20Rik |
RIKEN cDNA 6820431F20 gene |
85 |
0.98 |
| chr9_55937961_55938121 | 1.34 |
Scaper |
S phase cyclin A-associated protein in the ER |
60 |
0.71 |
| chr7_111082645_111082802 | 1.32 |
Eif4g2 |
eukaryotic translation initiation factor 4, gamma 2 |
104 |
0.95 |
| chr3_119783201_119783359 | 1.26 |
Ptbp2 |
polypyrimidine tract binding protein 2 |
6 |
0.97 |
| chrX_73967268_73967441 | 1.24 |
Hcfc1 |
host cell factor C1 |
997 |
0.39 |
| chr15_100682914_100683075 | 1.21 |
Cela1 |
chymotrypsin-like elastase family, member 1 |
125 |
0.93 |
| chr13_55714490_55714677 | 1.20 |
Txndc15 |
thioredoxin domain containing 15 |
67 |
0.96 |
| chrX_103493546_103493745 | 1.20 |
Jpx |
Jpx transcript, Xist activator (non-protein coding) |
64 |
0.93 |
| chr4_95397393_95397566 | 1.18 |
Gm29064 |
predicted gene 29064 |
5311 |
0.26 |
| chr11_72796017_72796182 | 1.18 |
Cyb5d2 |
cytochrome b5 domain containing 2 |
5 |
0.62 |
| chr1_171294738_171294910 | 1.17 |
Ufc1 |
ubiquitin-fold modifier conjugating enzyme 1 |
116 |
0.89 |
| chr7_128740333_128740636 | 1.15 |
Mcmbp |
minichromosome maintenance complex binding protein |
2 |
0.95 |
| chr10_111009930_111010085 | 1.15 |
Zdhhc17 |
zinc finger, DHHC domain containing 17 |
4 |
0.98 |
| chr11_98773925_98774148 | 1.12 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
1297 |
0.27 |
| chr14_51101004_51101293 | 1.10 |
Rnase4 |
ribonuclease, RNase A family 4 |
4789 |
0.09 |
| chr7_45016046_45016197 | 1.09 |
Scaf1 |
SR-related CTD-associated factor 1 |
10 |
0.91 |
| chr16_32202814_32202973 | 1.06 |
Bex6 |
brain expressed family member 6 |
16707 |
0.11 |
| chr3_95434221_95434414 | 1.06 |
Arnt |
aryl hydrocarbon receptor nuclear translocator |
71 |
0.94 |
| chr19_46979408_46979575 | 1.06 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
9923 |
0.15 |
| chr6_31398699_31398872 | 1.05 |
Mkln1os |
muskelin 1, intracellular mediator containing kelch motifs, opposite strand |
12 |
0.51 |
| chr7_19952685_19952933 | 1.05 |
Rpl7a-ps8 |
ribosomal protein L7A, pseudogene 8 |
974 |
0.29 |
| chr5_134176723_134177153 | 1.04 |
Rcc1l |
reculator of chromosome condensation 1 like |
164 |
0.82 |
| chr15_80133045_80133212 | 1.03 |
Tab1 |
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
1 |
0.96 |
| chr8_95825329_95825480 | 1.02 |
Sap18b |
Sin3-associated polypeptide 18B |
58 |
0.94 |
| chr3_135440220_135441003 | 1.02 |
Ube2d3 |
ubiquitin-conjugating enzyme E2D 3 |
1400 |
0.24 |
| chrX_151520462_151520649 | 1.02 |
Phf8 |
PHD finger protein 8 |
117 |
0.96 |
| chr2_164911250_164911413 | 1.01 |
Zfp335os |
zinc finger protein 335, opposite strand |
23 |
0.86 |
| chr3_69004802_69004994 | 1.01 |
Smc4 |
structural maintenance of chromosomes 4 |
72 |
0.79 |
| chr5_33936341_33936510 | 1.00 |
Nelfa |
negative elongation factor complex member A, Whsc2 |
12 |
0.96 |
| chr15_102203362_102203697 | 1.00 |
Csad |
cysteine sulfinic acid decarboxylase |
75 |
0.55 |
| chr6_71203524_71203686 | 0.99 |
Fabp1 |
fatty acid binding protein 1, liver |
3778 |
0.15 |
| chr3_85821665_85821854 | 0.99 |
Fam160a1 |
family with sequence similarity 160, member A1 |
4468 |
0.2 |
| chrX_142966510_142966805 | 0.99 |
Ammecr1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
71 |
0.98 |
| chr8_3418238_3418389 | 0.97 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
8387 |
0.15 |
| chr7_6728225_6728524 | 0.97 |
Peg3 |
paternally expressed 3 |
2045 |
0.16 |
| chr9_119894768_119894935 | 0.97 |
Gm2449 |
predicted gene 2449 |
6 |
0.5 |
| chr5_52783008_52783165 | 0.96 |
Zcchc4 |
zinc finger, CCHC domain containing 4 |
0 |
0.98 |
| chr11_74897009_74897183 | 0.96 |
Sgsm2 |
small G protein signaling modulator 2 |
36 |
0.93 |
| chr7_6728633_6728957 | 0.95 |
Peg3 |
paternally expressed 3 |
1624 |
0.18 |
| chr4_70410468_70410632 | 0.94 |
Cdk5rap2 |
CDK5 regulatory subunit associated protein 2 |
107 |
0.98 |
| chr18_39491869_39492036 | 0.94 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
651 |
0.8 |
| chr1_134955895_134956232 | 0.93 |
Ppp1r12b |
protein phosphatase 1, regulatory subunit 12B |
121 |
0.96 |
| chr1_93754778_93754978 | 0.93 |
Thap4 |
THAP domain containing 4 |
14 |
0.5 |
| chr15_10872191_10872614 | 0.93 |
Gm29742 |
predicted gene, 29742 |
45035 |
0.12 |
| chr15_81585297_81585458 | 0.92 |
Ep300 |
E1A binding protein p300 |
20 |
0.81 |
| chr4_130663318_130663476 | 0.92 |
Pum1 |
pumilio RNA-binding family member 1 |
9 |
0.98 |
| chr10_79854529_79854703 | 0.91 |
Ptbp1 |
polypyrimidine tract binding protein 1 |
11 |
0.8 |
| chr18_6490321_6490488 | 0.91 |
Mir1893 |
microRNA 1893 |
242 |
0.42 |
| chr14_26534922_26535085 | 0.89 |
Slmap |
sarcolemma associated protein |
72 |
0.96 |
| chr11_80383172_80383332 | 0.88 |
Zfp207 |
zinc finger protein 207 |
27 |
0.97 |
| chrX_99820125_99820276 | 0.87 |
Tmem28 |
transmembrane protein 28 |
821 |
0.7 |
| chr10_79908872_79909048 | 0.87 |
Med16 |
mediator complex subunit 16 |
37 |
0.71 |
| chr7_141328038_141328207 | 0.87 |
Tmem80 |
transmembrane protein 80 |
8 |
0.86 |
| chr9_90113472_90114008 | 0.86 |
Morf4l1 |
mortality factor 4 like 1 |
549 |
0.72 |
| chr19_61265459_61265626 | 0.86 |
Gm50357 |
predicted gene, 50357 |
7753 |
0.13 |
| chr9_21510164_21510327 | 0.84 |
Tmed1 |
transmembrane p24 trafficking protein 1 |
7 |
0.95 |
| chr1_171410933_171411111 | 0.84 |
Usf1 |
upstream transcription factor 1 |
291 |
0.78 |
| chr17_34031607_34031784 | 0.84 |
Slc39a7 |
solute carrier family 39 (zinc transporter), member 7 |
13 |
0.42 |
| chr7_25076896_25077240 | 0.83 |
Zfp574 |
zinc finger protein 574 |
137 |
0.93 |
| chr5_137510334_137510492 | 0.82 |
Gm8066 |
predicted gene 8066 |
160 |
0.87 |
| chr14_51063014_51063572 | 0.82 |
Rnase12 |
ribonuclease, RNase A family, 12 (non-active) |
5632 |
0.09 |
| chr17_32283998_32284149 | 0.82 |
Brd4 |
bromodomain containing 4 |
17 |
0.93 |
| chr12_25142257_25142418 | 0.82 |
Gm36287 |
predicted gene, 36287 |
8578 |
0.16 |
| chrX_73966318_73966576 | 0.82 |
Hcfc1 |
host cell factor C1 |
90 |
0.95 |
| chr19_43440308_43440459 | 0.82 |
Cnnm1 |
cyclin M1 |
53 |
0.52 |
| chr7_44895965_44896136 | 0.81 |
Fuz |
fuzzy planar cell polarity protein |
29 |
0.92 |
| chr6_115538998_115539388 | 0.81 |
Gm36355 |
predicted gene, 36355 |
2293 |
0.23 |
| chr9_83599585_83599736 | 0.80 |
Sh3bgrl2 |
SH3 domain binding glutamic acid-rich protein like 2 |
22829 |
0.16 |
| chr2_179976547_179976703 | 0.80 |
Taf4 |
TATA-box binding protein associated factor 4 |
21 |
0.75 |
| chr8_84729906_84730267 | 0.80 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
7079 |
0.11 |
| chr17_73845744_73845921 | 0.80 |
Gm4948 |
predicted gene 4948 |
25518 |
0.16 |
| chr12_80644107_80644265 | 0.80 |
Slc39a9 |
solute carrier family 39 (zinc transporter), member 9 |
40 |
0.52 |
| chr7_59228475_59228638 | 0.79 |
Ube3a |
ubiquitin protein ligase E3A |
194 |
0.83 |
| chr3_133544244_133544404 | 0.79 |
Tet2 |
tet methylcytosine dioxygenase 2 |
22 |
0.94 |
| chr10_59616602_59616753 | 0.79 |
Mcu |
mitochondrial calcium uniporter |
2 |
0.93 |
| chr2_117374449_117374636 | 0.79 |
Rasgrp1 |
RAS guanyl releasing protein 1 |
31541 |
0.18 |
| chr19_46761710_46761872 | 0.78 |
Cnnm2 |
cyclin M2 |
186 |
0.94 |
| chr16_84834907_84835088 | 0.78 |
Atp5j |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
8 |
0.54 |
| chr10_128821663_128821988 | 0.78 |
Sarnp |
SAP domain containing ribonucleoprotein |
69 |
0.64 |
| chr10_82763951_82764143 | 0.78 |
Nfyb |
nuclear transcription factor-Y beta |
54 |
0.96 |
| chr4_149977780_149977941 | 0.77 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
22817 |
0.13 |
| chr7_35028668_35028899 | 0.77 |
Pepd |
peptidase D |
11699 |
0.1 |
| chr4_154964009_154964172 | 0.76 |
Pank4 |
pantothenate kinase 4 |
33 |
0.95 |
| chr10_61372644_61372803 | 0.76 |
Gm48074 |
predicted gene, 48074 |
8325 |
0.12 |
| chr11_117654755_117654912 | 0.75 |
Tnrc6c |
trinucleotide repeat containing 6C |
20 |
0.98 |
| chrX_73966934_73967096 | 0.75 |
Hcfc1 |
host cell factor C1 |
658 |
0.56 |
| chr3_30602517_30602677 | 0.74 |
Mynn |
myoneurin |
34 |
0.96 |
| chr3_107332987_107333148 | 0.73 |
Rbm15 |
RNA binding motif protein 15 |
34 |
0.97 |
| chr1_161035012_161035182 | 0.73 |
Gas5 |
growth arrest specific 5 |
30 |
0.59 |
| chr5_150673686_150673840 | 0.73 |
Pds5b |
PDS5 cohesin associated factor B |
24 |
0.96 |
| chr1_156558681_156558856 | 0.72 |
Abl2 |
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
18 |
0.97 |
| chr11_109651050_109651235 | 0.72 |
Prkar1a |
protein kinase, cAMP dependent regulatory, type I, alpha |
221 |
0.92 |
| chr7_127512290_127512459 | 0.72 |
Srcap |
Snf2-related CREBBP activator protein |
73 |
0.83 |
| chr5_150665558_150665899 | 0.72 |
N4bp2l2 |
NEDD4 binding protein 2-like 2 |
116 |
0.94 |
| chr6_35503019_35503170 | 0.72 |
Gm23053 |
predicted gene, 23053 |
21544 |
0.21 |
| chr17_45506742_45506903 | 0.71 |
Aars2 |
alanyl-tRNA synthetase 2, mitochondrial |
19 |
0.96 |
| chr2_180710267_180710418 | 0.71 |
Gid8 |
GID complex subunit 8 |
225 |
0.61 |
| chr8_69625483_69625772 | 0.71 |
Zfp868 |
zinc finger protein 868 |
79 |
0.96 |
| chr7_19496130_19496424 | 0.71 |
Exoc3l2 |
exocyst complex component 3-like 2 |
7221 |
0.08 |
| chrX_8074492_8074668 | 0.70 |
Suv39h1 |
suppressor of variegation 3-9 1 |
41 |
0.93 |
| chr18_61720939_61721101 | 0.70 |
Grpel2 |
GrpE-like 2, mitochondrial |
1798 |
0.23 |
| chr18_39491582_39491748 | 0.70 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
364 |
0.91 |
| chr15_58076166_58076331 | 0.70 |
Zhx1 |
zinc fingers and homeoboxes 1 |
3 |
0.51 |
| chr18_66002529_66002702 | 0.69 |
Lman1 |
lectin, mannose-binding, 1 |
3 |
0.97 |
| chr2_90940263_90940532 | 0.69 |
Celf1 |
CUGBP, Elav-like family member 1 |
0 |
0.96 |
| chr5_135187024_135187335 | 0.69 |
Baz1b |
bromodomain adjacent to zinc finger domain, 1B |
85 |
0.95 |
| chr12_83631971_83632288 | 0.69 |
Rbm25 |
RNA binding motif protein 25 |
123 |
0.94 |
| chr6_133105613_133106043 | 0.69 |
Smim10l1 |
small integral membrane protein 10 like 1 |
336 |
0.76 |
| chr12_112976741_112977039 | 0.69 |
Btbd6 |
BTB (POZ) domain containing 6 |
56 |
0.94 |
| chr16_87354115_87354279 | 0.69 |
N6amt1 |
N-6 adenine-specific DNA methyltransferase 1 (putative) |
12 |
0.98 |
| chr4_149099780_149099942 | 0.68 |
Pex14 |
peroxisomal biogenesis factor 14 |
15 |
0.96 |
| chr8_126539889_126540054 | 0.68 |
Gm26759 |
predicted gene, 26759 |
26999 |
0.18 |
| chr9_119322355_119322520 | 0.68 |
Oxsr1 |
oxidative-stress responsive 1 |
10 |
0.96 |
| chrX_73008216_73008393 | 0.68 |
Mir2137 |
microRNA 2137 |
16225 |
0.1 |
| chr8_71396533_71396900 | 0.68 |
Babam1 |
BRISC and BRCA1 A complex member 1 |
145 |
0.87 |
| chr3_121885017_121885175 | 0.68 |
Gm42593 |
predicted gene 42593 |
22254 |
0.15 |
| chr2_168154454_168154811 | 0.68 |
E130018N17Rik |
RIKEN cDNA E130018N17 gene |
119 |
0.92 |
| chr7_121803443_121803611 | 0.67 |
Gm44740 |
predicted gene 44740 |
35538 |
0.14 |
| chr6_4601091_4601254 | 0.67 |
Casd1 |
CAS1 domain containing 1 |
26 |
0.97 |
| chr9_108438992_108439149 | 0.67 |
BC048562 |
cDNA sequence BC048562 |
2576 |
0.1 |
| chr14_76414563_76414722 | 0.67 |
Tsc22d1 |
TSC22 domain family, member 1 |
319 |
0.93 |
| chr8_111259173_111259338 | 0.66 |
Glg1 |
golgi apparatus protein 1 |
39 |
0.97 |
| chr1_106200351_106200637 | 0.66 |
Gm38235 |
predicted gene, 38235 |
18820 |
0.17 |
| chrX_8132783_8132963 | 0.66 |
Wdr13 |
WD repeat domain 13 |
15 |
0.95 |
| chr17_56673236_56673398 | 0.66 |
Ranbp3 |
RAN binding protein 3 |
23 |
0.96 |
| chr13_36117789_36118096 | 0.66 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
62 |
0.86 |
| chr11_61408235_61408402 | 0.66 |
Gm45605 |
predicted gene 45605 |
300 |
0.85 |
| chr3_152714008_152714166 | 0.65 |
Pigk |
phosphatidylinositol glycan anchor biosynthesis, class K |
13 |
0.98 |
| chr6_126849087_126849242 | 0.65 |
Ndufa9 |
NADH:ubiquinone oxidoreductase subunit A9 |
28 |
0.96 |
| chr6_54816726_54816912 | 0.65 |
Znrf2 |
zinc and ring finger 2 |
97 |
0.98 |
| chr3_65957458_65957685 | 0.65 |
Ccnl1 |
cyclin L1 |
82 |
0.64 |
| chr16_90284402_90284553 | 0.64 |
Scaf4 |
SR-related CTD-associated factor 4 |
26 |
0.97 |
| chr18_24530179_24530342 | 0.63 |
Rprd1a |
regulation of nuclear pre-mRNA domain containing 1A |
56 |
0.97 |
| chr11_69672405_69672596 | 0.63 |
Eif4a1 |
eukaryotic translation initiation factor 4A1 |
77 |
0.86 |
| chr11_83753608_83753766 | 0.63 |
Heatr6 |
HEAT repeat containing 6 |
9 |
0.95 |
| chr6_34029353_34029536 | 0.62 |
Lrguk |
leucine-rich repeats and guanylate kinase domain containing |
4 |
0.98 |
| chr6_71823720_71824038 | 0.62 |
Mrpl35 |
mitochondrial ribosomal protein L35 |
69 |
0.95 |
| chr2_49451895_49452257 | 0.62 |
Epc2 |
enhancer of polycomb homolog 2 |
590 |
0.81 |
| chr3_51978070_51978267 | 0.61 |
Gm20089 |
predicted gene, 20089 |
2773 |
0.18 |
| chr1_62425224_62425405 | 0.61 |
Pard3bos3 |
par-3 family cell polarity regulator beta, opposite strand 3 |
73947 |
0.11 |
| chr7_6729499_6729657 | 0.61 |
Peg3 |
paternally expressed 3 |
841 |
0.29 |
| chr2_18677284_18677462 | 0.61 |
Bmi1 |
Bmi1 polycomb ring finger oncogene |
93 |
0.95 |
| chr11_116274095_116274258 | 0.60 |
Srp68 |
signal recognition particle 68 |
41 |
0.95 |
| chr6_30390971_30391133 | 0.60 |
Zc3hc1 |
zinc finger, C3HC type 1 |
24 |
0.97 |
| chr18_7869642_7869793 | 0.60 |
Wac |
WW domain containing adaptor with coiled-coil |
1 |
0.92 |
| chr9_64811222_64811386 | 0.60 |
Dennd4a |
DENN/MADD domain containing 4A |
36 |
0.98 |
| chrY_1286191_1286403 | 0.60 |
Ddx3y |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
285 |
0.88 |
| chr4_117343183_117343345 | 0.60 |
Rnf220 |
ring finger protein 220 |
32193 |
0.12 |
| chr14_54603440_54603604 | 0.60 |
4931414P19Rik |
RIKEN cDNA 4931414P19 gene |
2471 |
0.12 |
| chr18_43477736_43477891 | 0.59 |
Eif3j2 |
eukaryotic translation initiation factor 3, subunit J2 |
17 |
0.5 |
| chr1_88146304_88146455 | 0.59 |
Gm15369 |
predicted gene 15369 |
3 |
0.93 |
| chr9_22002881_22003040 | 0.59 |
Prkcsh |
protein kinase C substrate 80K-H |
58 |
0.79 |
| chr2_52072729_52072909 | 0.59 |
Rif1 |
replication timing regulatory factor 1 |
13 |
0.98 |
| chr13_96542699_96542853 | 0.59 |
Col4a3bp |
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
21 |
0.69 |
| chr17_12383296_12383451 | 0.59 |
Plg |
plasminogen |
4714 |
0.18 |
| chr1_171345565_171345724 | 0.58 |
Nit1 |
nitrilase 1 |
1 |
0.49 |
| chr18_38212801_38212957 | 0.58 |
2010320O07Rik |
RIKEN cDNA 2010320O07 gene |
825 |
0.35 |
| chr16_94569602_94569765 | 0.58 |
Dyrk1a |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a |
327 |
0.89 |
| chr1_105993956_105994124 | 0.58 |
Zcchc2 |
zinc finger, CCHC domain containing 2 |
3351 |
0.2 |
| chr13_55622890_55623046 | 0.57 |
Caml |
calcium modulating ligand |
37 |
0.95 |
| chrX_38564603_38564760 | 0.57 |
Cul4b |
cullin 4B |
19 |
0.98 |
| chr7_121707472_121707627 | 0.57 |
Usp31 |
ubiquitin specific peptidase 31 |
296 |
0.55 |
| chr3_97767978_97768139 | 0.57 |
Pde4dip |
phosphodiesterase 4D interacting protein (myomegalin) |
49 |
0.98 |
| chr3_30793238_30793395 | 0.57 |
Sec62 |
SEC62 homolog (S. cerevisiae) |
20 |
0.76 |
| chr4_132768097_132768266 | 0.57 |
Rpa2 |
replication protein A2 |
151 |
0.93 |
| chr12_108179709_108179877 | 0.57 |
Ccnk |
cyclin K |
55 |
0.89 |
| chr16_97497728_97497915 | 0.57 |
Gm24777 |
predicted gene, 24777 |
1201 |
0.36 |
| chr9_98184797_98185092 | 0.56 |
2610303G11Rik |
RIKEN cDNA 2610303G11 gene |
2209 |
0.4 |
| chr11_6292471_6292636 | 0.56 |
Ogdh |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
52 |
0.96 |
| chr3_157947121_157947282 | 0.56 |
Ankrd13c |
ankyrin repeat domain 13c |
38 |
0.97 |
| chr16_32686224_32686404 | 0.56 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
5500 |
0.15 |
| chrX_18163035_18163247 | 0.56 |
Kdm6a |
lysine (K)-specific demethylase 6A |
493 |
0.85 |
| chr3_123353519_123353753 | 0.56 |
Sec24d |
Sec24 related gene family, member D (S. cerevisiae) |
1189 |
0.44 |
| chr1_191062397_191063065 | 0.56 |
Tatdn3 |
TatD DNase domain containing 3 |
173 |
0.62 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0019042 | viral latency(GO:0019042) |
| 0.5 | 1.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 0.7 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.3 | 1.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.3 | 0.9 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.3 | 0.8 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 | 1.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 1.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 0.7 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 0.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 0.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 0.7 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 0.7 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 2.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 0.9 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 2.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.6 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.6 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.2 | 0.6 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.2 | 0.5 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.2 | 0.9 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 2.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 0.3 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 1.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.4 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 0.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.8 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 2.0 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.4 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.9 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.8 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 3.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.4 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.2 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.1 | 1.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.9 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.3 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.3 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) rRNA transport(GO:0051029) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.2 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 0.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.5 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 1.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.4 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 0.1 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.1 | 0.3 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.1 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.6 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.6 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.7 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.9 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.1 | 0.3 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 1.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 1.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.2 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.4 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 2.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.2 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.1 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 1.8 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.4 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 1.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.2 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.3 | GO:0010453 | regulation of cell fate commitment(GO:0010453) |
| 0.0 | 0.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 | 0.7 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 1.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.5 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.0 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:0090151 | establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.0 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.0 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.0 | GO:2000510 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.0 | 0.1 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.0 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.8 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.0 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.0 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.3 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 1.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 0.8 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 0.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 0.5 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.6 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.6 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 1.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 1.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 1.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 1.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.3 | 3.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 2.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 1.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.4 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.5 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.2 | 0.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.8 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.4 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0046428 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.1 | 1.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.4 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 3.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.5 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.9 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0047419 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 1.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 0.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.5 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME MITOTIC M M G1 PHASES | Genes involved in Mitotic M-M/G1 phases |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |