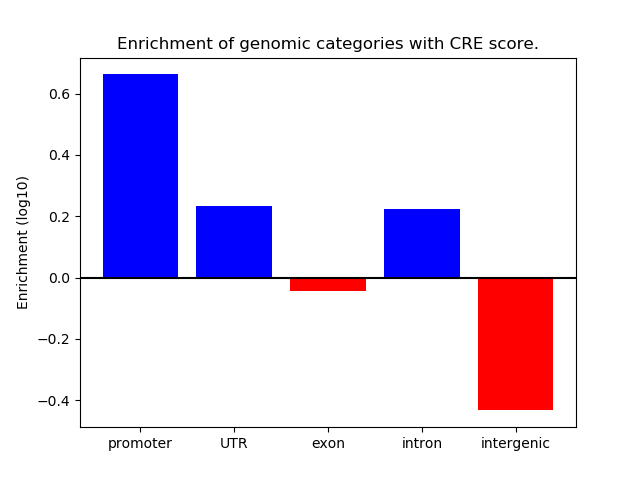
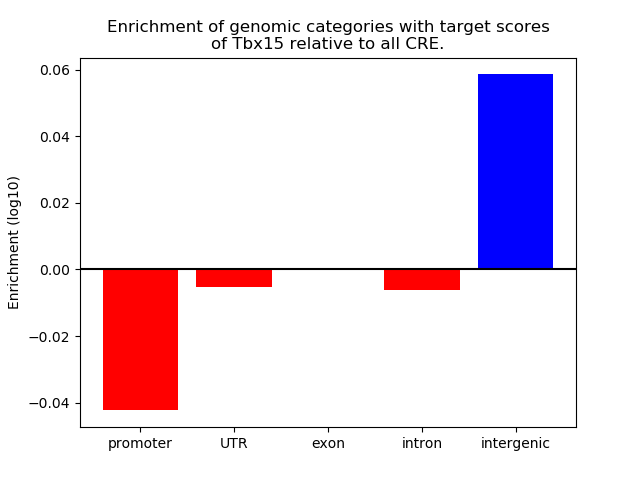
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
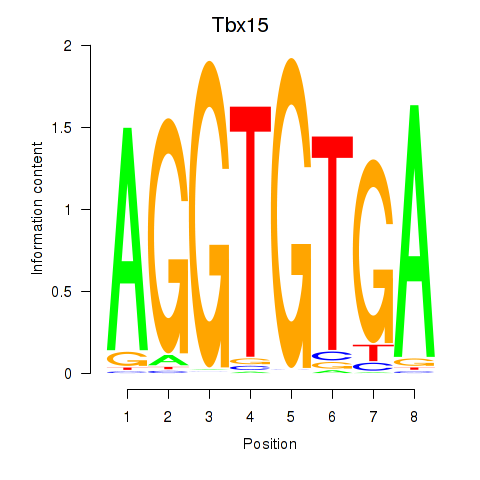
Results for Tbx15
Z-value: 0.39
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSMUSG00000027868.5 | T-box 15 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_99234096_99234265 | Tbx15 | 6201 | 0.153550 | -0.89 | 1.8e-02 | Click! |
Activity of the Tbx15 motif across conditions
Conditions sorted by the z-value of the Tbx15 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
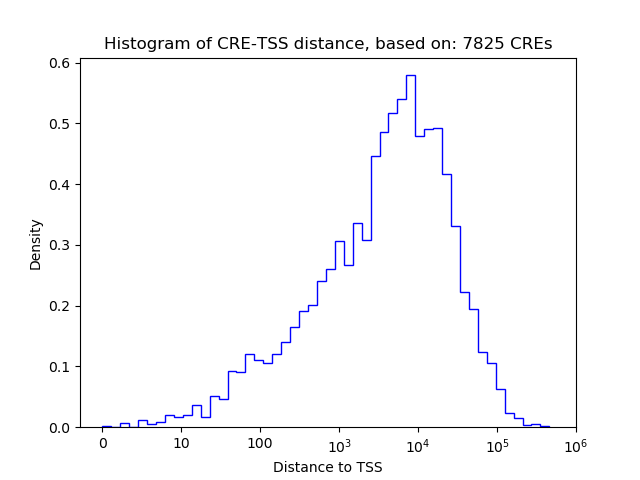
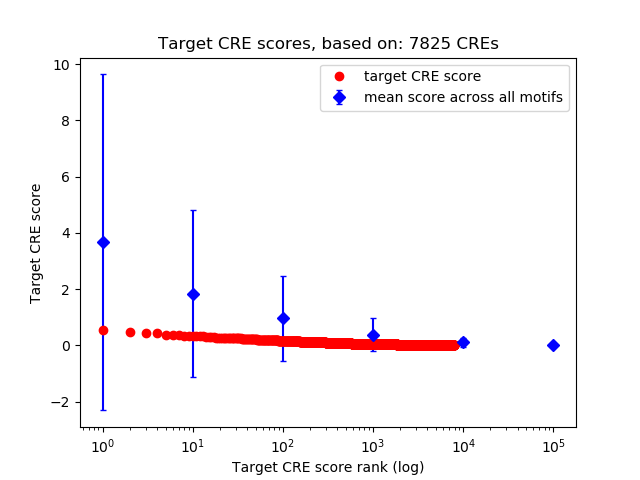
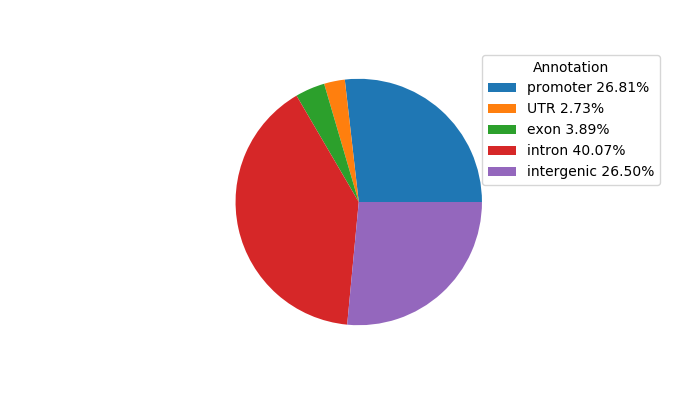
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_52388262_52388419 | 0.55 |
Gm23460 |
predicted gene, 23460 |
56204 |
0.11 |
| chr4_141316341_141316787 | 0.49 |
Epha2 |
Eph receptor A2 |
349 |
0.78 |
| chr4_128946455_128946665 | 0.44 |
Gm15904 |
predicted gene 15904 |
10555 |
0.15 |
| chr5_53437072_53437250 | 0.42 |
Gm26486 |
predicted gene, 26486 |
18588 |
0.17 |
| chr11_59640294_59640451 | 0.36 |
Mprip |
myosin phosphatase Rho interacting protein |
20933 |
0.11 |
| chr7_100658121_100658314 | 0.36 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
147 |
0.93 |
| chr4_11861132_11861344 | 0.35 |
Gm25002 |
predicted gene, 25002 |
87649 |
0.07 |
| chr5_145993052_145993431 | 0.35 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
1598 |
0.28 |
| chr14_65832955_65833117 | 0.34 |
Esco2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
872 |
0.57 |
| chr12_24463539_24463734 | 0.33 |
Gm16372 |
predicted pseudogene 16372 |
30020 |
0.14 |
| chr8_122174579_122174745 | 0.33 |
Zfp469 |
zinc finger protein 469 |
83958 |
0.07 |
| chr2_26405907_26406058 | 0.32 |
Inpp5e |
inositol polyphosphate-5-phosphatase E |
3145 |
0.11 |
| chr5_75046920_75047087 | 0.32 |
Gm18593 |
predicted gene, 18593 |
1476 |
0.27 |
| chr13_104615047_104615206 | 0.31 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
69805 |
0.12 |
| chr11_107226116_107226397 | 0.30 |
Gm11718 |
predicted gene 11718 |
35162 |
0.12 |
| chr10_78320688_78320839 | 0.29 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
73 |
0.94 |
| chr1_93135074_93135362 | 0.28 |
Agxt |
alanine-glyoxylate aminotransferase |
22 |
0.96 |
| chr12_80832436_80832620 | 0.28 |
Susd6 |
sushi domain containing 6 |
36180 |
0.11 |
| chr4_117116805_117116956 | 0.27 |
Ptch2 |
patched 2 |
3456 |
0.09 |
| chr4_134060274_134060433 | 0.27 |
Crybg2 |
crystallin beta-gamma domain containing 2 |
462 |
0.7 |
| chr5_119295643_119295794 | 0.26 |
n-R5s175 |
nuclear encoded rRNA 5S 175 |
1527 |
0.52 |
| chr5_117275916_117276216 | 0.26 |
Pebp1 |
phosphatidylethanolamine binding protein 1 |
11459 |
0.11 |
| chr5_125296722_125296910 | 0.26 |
Scarb1 |
scavenger receptor class B, member 1 |
2707 |
0.22 |
| chr14_20319722_20320198 | 0.26 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
8531 |
0.12 |
| chr12_21526957_21527150 | 0.26 |
Gm19196 |
predicted gene, 19196 |
16254 |
0.22 |
| chr7_104307089_104307261 | 0.26 |
Trim12a |
tripartite motif-containing 12A |
6141 |
0.08 |
| chr8_119416779_119416930 | 0.26 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
17270 |
0.13 |
| chr9_43225133_43225294 | 0.25 |
Oaf |
out at first homolog |
121 |
0.95 |
| chr2_126776033_126776199 | 0.25 |
Usp50 |
ubiquitin specific peptidase 50 |
5660 |
0.18 |
| chr7_104279565_104279731 | 0.25 |
Trim5 |
tripartite motif-containing 5 |
84 |
0.93 |
| chr6_56919800_56919965 | 0.25 |
Nt5c3 |
5'-nucleotidase, cytosolic III |
3842 |
0.15 |
| chr5_8157542_8157699 | 0.25 |
Gm21759 |
predicted gene, 21759 |
22016 |
0.16 |
| chr5_25019324_25019486 | 0.24 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
23652 |
0.16 |
| chr12_82914756_82914931 | 0.24 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
24312 |
0.21 |
| chr8_121201174_121201325 | 0.24 |
5033426O07Rik |
RIKEN cDNA 5033426O07 gene |
586 |
0.75 |
| chr13_80896624_80897028 | 0.24 |
Arrdc3 |
arrestin domain containing 3 |
6308 |
0.17 |
| chr17_80468453_80468787 | 0.24 |
Sos1 |
SOS Ras/Rac guanine nucleotide exchange factor 1 |
11349 |
0.21 |
| chr1_184093517_184093672 | 0.24 |
Dusp10 |
dual specificity phosphatase 10 |
59213 |
0.13 |
| chr2_181373148_181373303 | 0.23 |
Zgpat |
zinc finger, CCCH-type with G patch domain |
5169 |
0.1 |
| chr17_29093649_29093812 | 0.23 |
Cdkn1a |
cyclin-dependent kinase inhibitor 1A (P21) |
39 |
0.91 |
| chr1_182661161_182661312 | 0.23 |
Gm37516 |
predicted gene, 37516 |
18703 |
0.17 |
| chr8_120495609_120495777 | 0.22 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
7246 |
0.15 |
| chr11_86932862_86933027 | 0.22 |
Ypel2 |
yippee like 2 |
39080 |
0.14 |
| chr1_180757970_180758155 | 0.22 |
Gm37768 |
predicted gene, 37768 |
2226 |
0.19 |
| chr10_61337817_61337968 | 0.22 |
Pald1 |
phosphatase domain containing, paladin 1 |
4435 |
0.15 |
| chrX_150956676_150956837 | 0.22 |
Gm8424 |
predicted gene 8424 |
30597 |
0.13 |
| chr5_89416345_89416742 | 0.22 |
Gc |
vitamin D binding protein |
19085 |
0.24 |
| chr8_120244102_120244267 | 0.21 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
15728 |
0.17 |
| chr5_138076351_138076506 | 0.21 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
8656 |
0.09 |
| chr6_86636895_86637046 | 0.21 |
Asprv1 |
aspartic peptidase, retroviral-like 1 |
8806 |
0.11 |
| chr3_98311224_98311375 | 0.21 |
Phgdh |
3-phosphoglycerate dehydrogenase |
17314 |
0.14 |
| chr11_101372782_101373038 | 0.20 |
G6pc |
glucose-6-phosphatase, catalytic |
5349 |
0.06 |
| chr4_41637175_41637517 | 0.20 |
Gm12406 |
predicted gene 12406 |
2032 |
0.17 |
| chr14_55079684_55079867 | 0.20 |
Zfhx2 |
zinc finger homeobox 2 |
677 |
0.47 |
| chr14_30906842_30907058 | 0.20 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
7179 |
0.11 |
| chr1_133073713_133073881 | 0.20 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
5218 |
0.16 |
| chr2_29437237_29437388 | 0.20 |
Gm24976 |
predicted gene, 24976 |
21882 |
0.15 |
| chr2_3771544_3771776 | 0.20 |
Fam107b |
family with sequence similarity 107, member B |
952 |
0.57 |
| chr3_83996881_83997402 | 0.20 |
Tmem131l |
transmembrane 131 like |
28913 |
0.2 |
| chr1_132299853_132300009 | 0.20 |
Klhdc8a |
kelch domain containing 8A |
1305 |
0.31 |
| chr19_23066702_23066853 | 0.19 |
Gm50136 |
predicted gene, 50136 |
5304 |
0.21 |
| chr13_94708250_94708413 | 0.19 |
Gm32351 |
predicted gene, 32351 |
5613 |
0.17 |
| chr15_34510659_34511041 | 0.19 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
943 |
0.43 |
| chr7_72306635_72306827 | 0.19 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
123 |
0.98 |
| chr11_116209384_116209702 | 0.19 |
Ten1 |
TEN1 telomerase capping complex subunit |
1391 |
0.24 |
| chr14_74853189_74853575 | 0.19 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
31873 |
0.18 |
| chr10_71183397_71183774 | 0.19 |
C730027H18Rik |
RIKEN cDNA C730027H18 gene |
1533 |
0.26 |
| chr5_117997853_117998004 | 0.19 |
Fbxo21 |
F-box protein 21 |
18604 |
0.13 |
| chr3_95652051_95652202 | 0.19 |
Mcl1 |
myeloid cell leukemia sequence 1 |
6662 |
0.1 |
| chr3_27905130_27905320 | 0.19 |
Tmem212 |
transmembrane protein 212 |
8857 |
0.22 |
| chr13_44302017_44302180 | 0.19 |
Gm29676 |
predicted gene, 29676 |
21649 |
0.18 |
| chr6_89213939_89214095 | 0.19 |
Gm839 |
predicted gene 839 |
2220 |
0.22 |
| chr10_61480037_61480223 | 0.19 |
D830039M14Rik |
RIKEN cDNA D830039M14 gene |
3193 |
0.13 |
| chr1_155112239_155112390 | 0.19 |
Ier5 |
immediate early response 5 |
12678 |
0.14 |
| chr15_25663250_25663426 | 0.19 |
Myo10 |
myosin X |
11172 |
0.18 |
| chr13_91765048_91765525 | 0.19 |
Gm27656 |
predicted gene, 27656 |
13180 |
0.14 |
| chr5_136965885_136966036 | 0.18 |
Cldn15 |
claudin 15 |
656 |
0.49 |
| chr12_84203864_84204015 | 0.18 |
Gm31513 |
predicted gene, 31513 |
7970 |
0.11 |
| chr19_3687091_3687242 | 0.18 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
602 |
0.61 |
| chr19_3329840_3329991 | 0.18 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
3810 |
0.15 |
| chr10_30810298_30810917 | 0.18 |
Ncoa7 |
nuclear receptor coactivator 7 |
7281 |
0.16 |
| chr19_7456855_7457030 | 0.18 |
Rtn3 |
reticulon 3 |
26260 |
0.1 |
| chr8_103538740_103538926 | 0.18 |
Gm45277 |
predicted gene 45277 |
24790 |
0.23 |
| chr5_135139962_135140114 | 0.18 |
Mlxipl |
MLX interacting protein-like |
6407 |
0.11 |
| chr4_116695063_116695819 | 0.18 |
Prdx1 |
peroxiredoxin 1 |
8129 |
0.1 |
| chr10_28178649_28178847 | 0.18 |
Gm22370 |
predicted gene, 22370 |
35373 |
0.21 |
| chr8_10857032_10857204 | 0.18 |
Gm32540 |
predicted gene, 32540 |
9068 |
0.12 |
| chr5_66041014_66041178 | 0.18 |
Rbm47 |
RNA binding motif protein 47 |
13456 |
0.12 |
| chr5_139197343_139197508 | 0.17 |
Sun1 |
Sad1 and UNC84 domain containing 1 |
3212 |
0.18 |
| chr17_86754051_86754206 | 0.17 |
Epas1 |
endothelial PAS domain protein 1 |
428 |
0.83 |
| chr5_134983246_134983642 | 0.17 |
Cldn3 |
claudin 3 |
2770 |
0.11 |
| chr1_184875101_184875316 | 0.17 |
C130074G19Rik |
RIKEN cDNA C130074G19 gene |
8010 |
0.17 |
| chr9_60926310_60926619 | 0.17 |
Gm47923 |
predicted gene, 47923 |
7241 |
0.21 |
| chr10_25621100_25621500 | 0.17 |
Gm36908 |
predicted gene, 36908 |
263 |
0.92 |
| chr18_6208229_6208380 | 0.17 |
Gm17036 |
predicted gene 17036 |
5609 |
0.19 |
| chr11_117872061_117872251 | 0.17 |
Tha1 |
threonine aldolase 1 |
245 |
0.79 |
| chr8_128465325_128465813 | 0.16 |
Nrp1 |
neuropilin 1 |
106172 |
0.06 |
| chr1_106309746_106309897 | 0.16 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
7417 |
0.25 |
| chr2_128174660_128174970 | 0.16 |
Gm14009 |
predicted gene 14009 |
15172 |
0.2 |
| chr11_79230285_79230459 | 0.16 |
Wsb1 |
WD repeat and SOCS box-containing 1 |
12577 |
0.16 |
| chr5_107837254_107837906 | 0.16 |
Evi5 |
ecotropic viral integration site 5 |
100 |
0.94 |
| chr14_51105673_51105825 | 0.16 |
Eddm3b |
epididymal protein 3B |
8677 |
0.08 |
| chr13_64361503_64361746 | 0.16 |
Gm49230 |
predicted gene, 49230 |
113 |
0.86 |
| chr7_118244089_118244267 | 0.16 |
4930583K01Rik |
RIKEN cDNA 4930583K01 gene |
313 |
0.63 |
| chr9_43737905_43738092 | 0.16 |
Gm30015 |
predicted gene, 30015 |
1043 |
0.39 |
| chr17_56615160_56615323 | 0.16 |
Rpl36 |
ribosomal protein L36 |
1825 |
0.19 |
| chr3_37902815_37902966 | 0.16 |
Gm20755 |
predicted gene, 20755 |
4077 |
0.21 |
| chr14_118124260_118124415 | 0.16 |
Tgds |
TDP-glucose 4,6-dehydratase |
8407 |
0.18 |
| chr19_40499284_40499488 | 0.16 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
11640 |
0.19 |
| chr1_167365951_167366119 | 0.16 |
Aldh9a1 |
aldehyde dehydrogenase 9, subfamily A1 |
1869 |
0.25 |
| chr1_191822042_191822193 | 0.16 |
Nek2 |
NIMA (never in mitosis gene a)-related expressed kinase 2 |
65 |
0.58 |
| chr6_134614784_134614949 | 0.16 |
Gm38910 |
predicted gene, 38910 |
7185 |
0.14 |
| chr2_30363854_30364005 | 0.16 |
Miga2 |
mitoguardin 2 |
304 |
0.79 |
| chr6_71203783_71204000 | 0.16 |
Fabp1 |
fatty acid binding protein 1, liver |
4064 |
0.15 |
| chrX_73466714_73466865 | 0.16 |
Haus7 |
HAUS augmin-like complex, subunit 7 |
7745 |
0.12 |
| chr11_50199089_50199272 | 0.16 |
Sqstm1 |
sequestosome 1 |
3774 |
0.13 |
| chr3_85813449_85813618 | 0.16 |
Fam160a1 |
family with sequence similarity 160, member A1 |
3758 |
0.23 |
| chr2_168207294_168207658 | 0.15 |
Adnp |
activity-dependent neuroprotective protein |
364 |
0.77 |
| chr2_72866286_72866437 | 0.15 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
52644 |
0.12 |
| chr4_8755441_8755604 | 0.15 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
4084 |
0.33 |
| chr10_79855965_79856127 | 0.15 |
Ptbp1 |
polypyrimidine tract binding protein 1 |
1008 |
0.2 |
| chr10_7832157_7832704 | 0.15 |
Zc3h12d |
zinc finger CCCH type containing 12D |
40 |
0.96 |
| chr11_103011230_103011391 | 0.15 |
Dcakd |
dephospho-CoA kinase domain containing |
5858 |
0.13 |
| chr9_91193419_91193605 | 0.15 |
Gm29602 |
predicted gene 29602 |
22503 |
0.17 |
| chr1_155888091_155888242 | 0.15 |
Cep350 |
centrosomal protein 350 |
2152 |
0.22 |
| chr7_84135967_84136134 | 0.15 |
Abhd17c |
abhydrolase domain containing 17C |
11765 |
0.15 |
| chr4_99201116_99201393 | 0.15 |
Atg4c |
autophagy related 4C, cysteine peptidase |
7107 |
0.19 |
| chr5_143412330_143412481 | 0.15 |
Kdelr2 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
8567 |
0.08 |
| chr8_70680106_70680375 | 0.15 |
Lsm4 |
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
2575 |
0.12 |
| chr10_120977866_120978049 | 0.15 |
Lemd3 |
LEM domain containing 3 |
1375 |
0.33 |
| chr3_98297519_98297734 | 0.15 |
Gm43189 |
predicted gene 43189 |
10638 |
0.14 |
| chr15_75996647_75996807 | 0.15 |
Mapk15 |
mitogen-activated protein kinase 15 |
14 |
0.94 |
| chr8_91379183_91379600 | 0.15 |
Fto |
fat mass and obesity associated |
11164 |
0.15 |
| chr1_58475799_58475950 | 0.15 |
Orc2 |
origin recognition complex, subunit 2 |
5575 |
0.13 |
| chr17_80483283_80483457 | 0.15 |
Sos1 |
SOS Ras/Rac guanine nucleotide exchange factor 1 |
2917 |
0.29 |
| chr4_115815775_115815933 | 0.15 |
Mob3c |
MOB kinase activator 3C |
12238 |
0.12 |
| chr2_105901718_105901869 | 0.14 |
Elp4 |
elongator acetyltransferase complex subunit 4 |
2690 |
0.24 |
| chr4_116708269_116708638 | 0.14 |
Mmachc |
methylmalonic aciduria cblC type, with homocystinuria |
47 |
0.5 |
| chr4_107159322_107159488 | 0.14 |
Tceanc2 |
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
18263 |
0.12 |
| chr5_102599017_102599403 | 0.14 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
115921 |
0.06 |
| chr5_66139803_66139965 | 0.14 |
Rbm47 |
RNA binding motif protein 47 |
11072 |
0.11 |
| chr11_53420114_53420567 | 0.14 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
2830 |
0.12 |
| chr5_124092063_124092447 | 0.14 |
Abcb9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
1993 |
0.19 |
| chr11_79670395_79670546 | 0.14 |
Rab11fip4os2 |
RAB11 family interacting protein 4 (class II), opposite strand 2 |
4613 |
0.13 |
| chr1_181010068_181010284 | 0.14 |
Ephx1 |
epoxide hydrolase 1, microsomal |
7393 |
0.09 |
| chr14_47296314_47296470 | 0.14 |
Mapk1ip1l |
mitogen-activated protein kinase 1 interacting protein 1-like |
1874 |
0.15 |
| chr9_24378970_24379135 | 0.14 |
Gm22380 |
predicted gene, 22380 |
27934 |
0.15 |
| chr15_102181335_102181499 | 0.14 |
Csad |
cysteine sulfinic acid decarboxylase |
1556 |
0.24 |
| chr19_4149325_4149619 | 0.14 |
Coro1b |
coronin, actin binding protein 1B |
805 |
0.28 |
| chr5_112281749_112281914 | 0.14 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
5124 |
0.13 |
| chr11_106581287_106581448 | 0.14 |
Tex2 |
testis expressed gene 2 |
767 |
0.66 |
| chr13_32824374_32824659 | 0.14 |
Wrnip1 |
Werner helicase interacting protein 1 |
3750 |
0.17 |
| chr9_63613110_63613389 | 0.14 |
Aagab |
alpha- and gamma-adaptin binding protein |
3739 |
0.23 |
| chr1_171454993_171455144 | 0.14 |
F11r |
F11 receptor |
7782 |
0.1 |
| chr8_75103139_75103449 | 0.14 |
Mcm5 |
minichromosome maintenance complex component 5 |
6275 |
0.14 |
| chr10_19976391_19976640 | 0.13 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
25460 |
0.2 |
| chr7_112187666_112187837 | 0.13 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
28694 |
0.2 |
| chr7_110840147_110840515 | 0.13 |
Rnf141 |
ring finger protein 141 |
2802 |
0.22 |
| chr14_57259417_57259587 | 0.13 |
Gm8996 |
predicted gene 8996 |
42269 |
0.13 |
| chr6_58879817_58880006 | 0.13 |
Herc3 |
hect domain and RLD 3 |
5568 |
0.21 |
| chr1_45922514_45922876 | 0.13 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
614 |
0.65 |
| chr3_85974604_85974787 | 0.13 |
Sh3d19 |
SH3 domain protein D19 |
3586 |
0.14 |
| chr7_48892680_48892953 | 0.13 |
Gm2788 |
predicted gene 2788 |
5586 |
0.14 |
| chr6_15893625_15894040 | 0.13 |
Gm43990 |
predicted gene, 43990 |
57743 |
0.15 |
| chr11_118265609_118265912 | 0.13 |
Usp36 |
ubiquitin specific peptidase 36 |
288 |
0.89 |
| chr2_31558979_31559141 | 0.13 |
Gm13426 |
predicted gene 13426 |
6365 |
0.15 |
| chr7_80360672_80360826 | 0.13 |
Man2a2 |
mannosidase 2, alpha 2 |
838 |
0.42 |
| chr11_109500636_109500796 | 0.13 |
Gm22378 |
predicted gene, 22378 |
892 |
0.5 |
| chr1_82239925_82240111 | 0.13 |
Gm9747 |
predicted gene 9747 |
6906 |
0.23 |
| chr8_119417171_119417322 | 0.13 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
16878 |
0.13 |
| chr6_146473719_146473882 | 0.13 |
Gm15720 |
predicted gene 15720 |
10193 |
0.19 |
| chr5_114435733_114435884 | 0.13 |
Mmab |
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human) |
8184 |
0.14 |
| chr7_19228690_19228852 | 0.13 |
Opa3 |
optic atrophy 3 |
388 |
0.68 |
| chr1_74304286_74304581 | 0.13 |
Tmbim1 |
transmembrane BAX inhibitor motif containing 1 |
28 |
0.94 |
| chr3_115815549_115815774 | 0.13 |
Dph5 |
diphthamide biosynthesis 5 |
72176 |
0.07 |
| chr10_83032672_83032849 | 0.13 |
Gm10773 |
predicted gene 10773 |
1065 |
0.55 |
| chr11_109488275_109488451 | 0.13 |
Arsg |
arylsulfatase G |
2757 |
0.18 |
| chr8_41545072_41545241 | 0.13 |
Gapdh-ps14 |
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 14 |
64513 |
0.13 |
| chr7_99694133_99694291 | 0.13 |
Gm34280 |
predicted gene, 34280 |
85 |
0.93 |
| chr10_121486376_121486686 | 0.13 |
Gm40787 |
predicted gene, 40787 |
2455 |
0.18 |
| chr8_13037092_13037395 | 0.13 |
F10 |
coagulation factor X |
65 |
0.94 |
| chr12_8676206_8676575 | 0.13 |
Pum2 |
pumilio RNA-binding family member 2 |
1144 |
0.56 |
| chr3_100201640_100202182 | 0.13 |
Gdap2 |
ganglioside-induced differentiation-associated-protein 2 |
1224 |
0.52 |
| chr8_46837465_46837662 | 0.13 |
Gm45481 |
predicted gene 45481 |
3651 |
0.22 |
| chr5_136526310_136526483 | 0.13 |
Gm38082 |
predicted gene, 38082 |
10081 |
0.22 |
| chr8_94172420_94173095 | 0.13 |
Mt2 |
metallothionein 2 |
93 |
0.88 |
| chr13_58123386_58123686 | 0.13 |
Hnrnpa0 |
heterogeneous nuclear ribonucleoprotein A0 |
5020 |
0.14 |
| chr5_145743023_145743196 | 0.13 |
Gm43626 |
predicted gene 43626 |
17436 |
0.15 |
| chr1_58097131_58097333 | 0.13 |
Aox3 |
aldehyde oxidase 3 |
15898 |
0.18 |
| chr6_113434799_113434964 | 0.13 |
Cidec |
cell death-inducing DFFA-like effector c |
117 |
0.9 |
| chr8_72222949_72223319 | 0.12 |
Fam32a |
family with sequence similarity 32, member A |
3370 |
0.12 |
| chr11_21398793_21399367 | 0.12 |
Ugp2 |
UDP-glucose pyrophosphorylase 2 |
27879 |
0.12 |
| chr18_31938569_31938916 | 0.12 |
Lims2 |
LIM and senescent cell antigen like domains 2 |
676 |
0.61 |
| chr3_95643397_95643587 | 0.12 |
E330034L11Rik |
RIKEN cDNA E330034L11 gene |
4188 |
0.11 |
| chr13_47179143_47179300 | 0.12 |
Rnf144b |
ring finger protein 144B |
14604 |
0.15 |
| chr8_25840737_25841255 | 0.12 |
Ash2l |
ASH2 like histone lysine methyltransferase complex subunit |
209 |
0.91 |
| chr16_30051044_30051883 | 0.12 |
9030404E10Rik |
RIKEN cDNA 9030404E10 gene |
11984 |
0.14 |
| chr19_43971247_43971535 | 0.12 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr1_134344955_134345133 | 0.12 |
Tmem183a |
transmembrane protein 183A |
5261 |
0.14 |
| chr11_55468108_55468290 | 0.12 |
G3bp1 |
GTPase activating protein (SH3 domain) binding protein 1 |
1486 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.2 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1902267 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0003164 | His-Purkinje system development(GO:0003164) bundle of His development(GO:0003166) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.0 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:1904833 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |