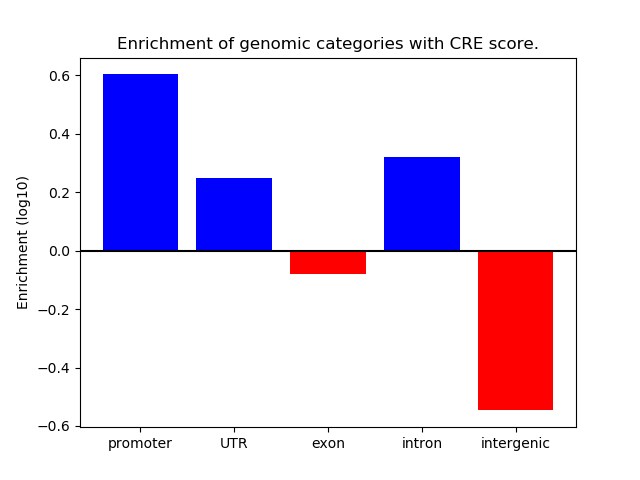
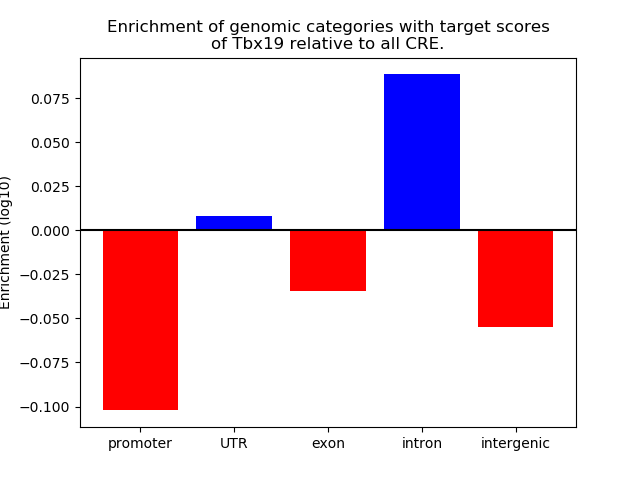
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
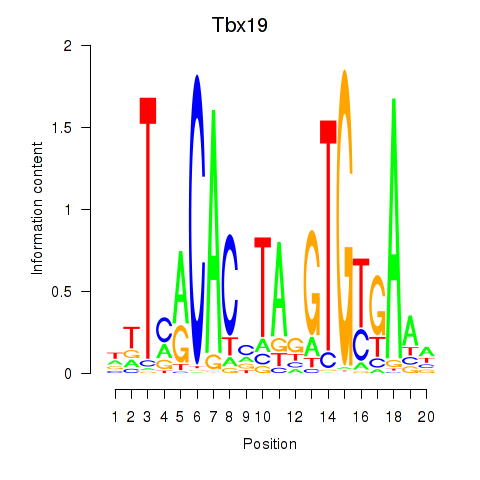
Results for Tbx19
Z-value: 1.75
Transcription factors associated with Tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx19
|
ENSMUSG00000026572.5 | T-box 19 |
Activity of the Tbx19 motif across conditions
Conditions sorted by the z-value of the Tbx19 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
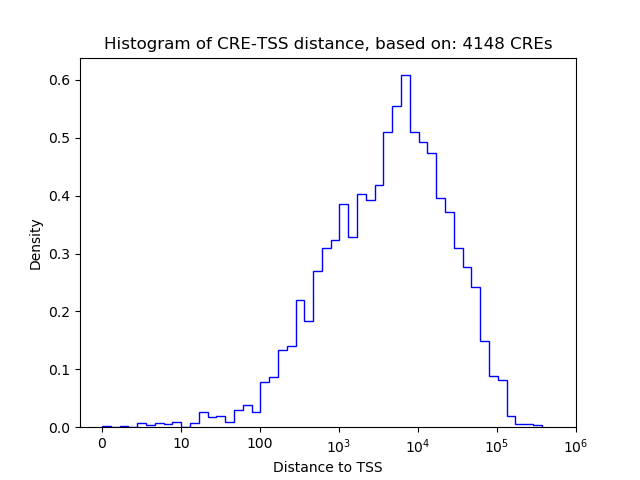
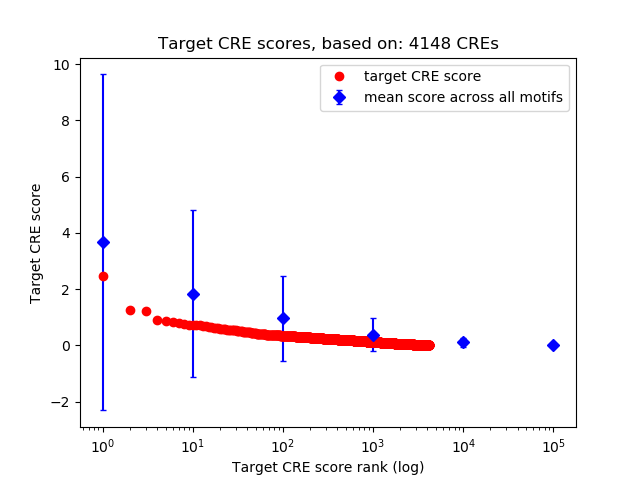
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_34500771_34501210 | 2.47 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
5667 |
0.12 |
| chr15_34501595_34501764 | 1.27 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6356 |
0.12 |
| chr15_34501830_34501985 | 1.22 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6584 |
0.12 |
| chr7_97415349_97415810 | 0.91 |
Thrsp |
thyroid hormone responsive |
1940 |
0.23 |
| chr5_105730043_105730194 | 0.85 |
Lrrc8d |
leucine rich repeat containing 8D |
1271 |
0.51 |
| chr6_24610042_24610446 | 0.84 |
Lmod2 |
leiomodin 2 (cardiac) |
12482 |
0.14 |
| chr19_44402814_44402965 | 0.81 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
3801 |
0.18 |
| chr8_36265077_36265268 | 0.77 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
15656 |
0.2 |
| chr17_34107373_34107524 | 0.73 |
Brd2 |
bromodomain containing 2 |
6284 |
0.05 |
| chr13_60479839_60480430 | 0.73 |
Gm48500 |
predicted gene, 48500 |
1164 |
0.45 |
| chr3_51230122_51230437 | 0.71 |
Gm38357 |
predicted gene, 38357 |
1638 |
0.29 |
| chr1_21266470_21266781 | 0.71 |
Gm28836 |
predicted gene 28836 |
4968 |
0.12 |
| chr7_66801636_66801787 | 0.69 |
Cers3 |
ceramide synthase 3 |
37542 |
0.13 |
| chr14_21422563_21422878 | 0.68 |
Gm25864 |
predicted gene, 25864 |
27754 |
0.17 |
| chr16_18340292_18340462 | 0.65 |
Tango2 |
transport and golgi organization 2 |
1299 |
0.25 |
| chr5_130026986_130027142 | 0.63 |
Asl |
argininosuccinate lyase |
2045 |
0.2 |
| chr15_34501373_34501524 | 0.61 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6125 |
0.12 |
| chr2_91195754_91195905 | 0.60 |
Nr1h3 |
nuclear receptor subfamily 1, group H, member 3 |
678 |
0.51 |
| chr5_24961125_24961358 | 0.60 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
24601 |
0.15 |
| chr13_107763794_107763945 | 0.59 |
Zswim6 |
zinc finger SWIM-type containing 6 |
15587 |
0.23 |
| chr10_15746557_15746708 | 0.59 |
Gm32283 |
predicted gene, 32283 |
7810 |
0.18 |
| chr11_4905764_4906019 | 0.57 |
Thoc5 |
THO complex 5 |
3679 |
0.15 |
| chr8_122047070_122047266 | 0.56 |
Banp |
BTG3 associated nuclear protein |
46321 |
0.11 |
| chr8_36290613_36290918 | 0.55 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
41249 |
0.14 |
| chr2_31501565_31501761 | 0.55 |
Ass1 |
argininosuccinate synthetase 1 |
937 |
0.56 |
| chr1_67187795_67187968 | 0.55 |
Gm15668 |
predicted gene 15668 |
61319 |
0.11 |
| chr9_109966945_109967106 | 0.54 |
Map4 |
microtubule-associated protein 4 |
1992 |
0.21 |
| chr2_6209909_6210643 | 0.54 |
Echdc3 |
enoyl Coenzyme A hydratase domain containing 3 |
2703 |
0.24 |
| chr13_93618232_93618425 | 0.54 |
Gm15622 |
predicted gene 15622 |
7054 |
0.17 |
| chr6_90472539_90472832 | 0.53 |
Klf15 |
Kruppel-like factor 15 |
5567 |
0.11 |
| chr19_34524558_34524859 | 0.51 |
Lipa |
lysosomal acid lipase A |
2703 |
0.19 |
| chr7_16526304_16526474 | 0.51 |
Gm17981 |
predicted gene, 17981 |
11251 |
0.12 |
| chr15_81247450_81247602 | 0.51 |
8430426J06Rik |
RIKEN cDNA 8430426J06 gene |
362 |
0.83 |
| chr11_98755184_98755335 | 0.50 |
Thra |
thyroid hormone receptor alpha |
1673 |
0.21 |
| chr19_44393747_44393961 | 0.50 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12836 |
0.14 |
| chr7_84129291_84129727 | 0.49 |
Abhd17c |
abhydrolase domain containing 17C |
18306 |
0.14 |
| chr15_95835869_95836082 | 0.48 |
Gm17546 |
predicted gene, 17546 |
5903 |
0.16 |
| chr1_63122681_63122832 | 0.48 |
Gm20342 |
predicted gene, 20342 |
5638 |
0.1 |
| chr1_181468293_181468488 | 0.47 |
Gm16539 |
predicted gene 16539 |
35990 |
0.14 |
| chr1_67197384_67197535 | 0.47 |
Gm15668 |
predicted gene 15668 |
51741 |
0.13 |
| chr15_52563909_52564070 | 0.46 |
Gm23267 |
predicted gene, 23267 |
64333 |
0.11 |
| chr10_40079067_40079342 | 0.46 |
Slc16a10 |
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
2599 |
0.21 |
| chr6_50773105_50773295 | 0.46 |
C530044C16Rik |
RIKEN cDNA C530044C16 gene |
2915 |
0.22 |
| chr9_42769307_42769458 | 0.45 |
Grik4 |
glutamate receptor, ionotropic, kainate 4 |
39317 |
0.19 |
| chr5_21795907_21796066 | 0.44 |
Psmc2 |
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
1883 |
0.25 |
| chr6_108454056_108454259 | 0.43 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
4546 |
0.21 |
| chr4_33467139_33467300 | 0.43 |
Gm11935 |
predicted gene 11935 |
14330 |
0.21 |
| chr1_131548025_131548226 | 0.43 |
Fam72a |
family with sequence similarity 72, member A |
20222 |
0.13 |
| chr1_67213641_67214357 | 0.43 |
Gm15668 |
predicted gene 15668 |
35201 |
0.17 |
| chr1_133694912_133695208 | 0.42 |
Lax1 |
lymphocyte transmembrane adaptor 1 |
4952 |
0.15 |
| chr7_19684170_19684408 | 0.42 |
Apoc4 |
apolipoprotein C-IV |
2810 |
0.1 |
| chr11_118454196_118454347 | 0.41 |
Gm11747 |
predicted gene 11747 |
724 |
0.56 |
| chr14_31635979_31636159 | 0.41 |
Gm49387 |
predicted gene, 49387 |
4809 |
0.15 |
| chr19_44398143_44398357 | 0.40 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
8440 |
0.15 |
| chr4_139519783_139519977 | 0.40 |
Iffo2 |
intermediate filament family orphan 2 |
10668 |
0.18 |
| chr3_58427270_58427444 | 0.40 |
Tsc22d2 |
TSC22 domain family, member 2 |
9873 |
0.19 |
| chr1_13670836_13671022 | 0.39 |
Xkr9 |
X-linked Kx blood group related 9 |
2158 |
0.29 |
| chr5_22587487_22587999 | 0.39 |
Gm9057 |
predicted gene 9057 |
23786 |
0.11 |
| chr1_88269284_88269537 | 0.39 |
Hjurp |
Holliday junction recognition protein |
18 |
0.96 |
| chr6_144071228_144071403 | 0.39 |
Sox5os1 |
SRY (sex determining region Y)-box 5, opposite strand 1 |
49407 |
0.17 |
| chr11_102152719_102152892 | 0.39 |
Tmem101 |
transmembrane protein 101 |
3542 |
0.1 |
| chr6_148209143_148209294 | 0.39 |
Ergic2 |
ERGIC and golgi 2 |
2759 |
0.21 |
| chr3_18241613_18241838 | 0.39 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
1613 |
0.45 |
| chr14_7827742_7828144 | 0.39 |
Flnb |
filamin, beta |
9986 |
0.16 |
| chr4_45443336_45443487 | 0.38 |
Slc25a51 |
solute carrier family 25, member 51 |
34645 |
0.11 |
| chr7_94427849_94428131 | 0.38 |
Gm44602 |
predicted gene 44602 |
47590 |
0.17 |
| chr4_45481507_45481658 | 0.38 |
Shb |
src homology 2 domain-containing transforming protein B |
1220 |
0.43 |
| chr14_61438447_61438598 | 0.38 |
Kpna3 |
karyopherin (importin) alpha 3 |
1352 |
0.34 |
| chr8_35418297_35418448 | 0.37 |
Gm45301 |
predicted gene 45301 |
8866 |
0.16 |
| chr10_24835678_24835882 | 0.37 |
Enpp3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
333 |
0.88 |
| chr11_106055229_106055380 | 0.37 |
Gm11646 |
predicted gene 11646 |
3452 |
0.13 |
| chr1_9797489_9798048 | 0.37 |
Sgk3 |
serum/glucocorticoid regulated kinase 3 |
339 |
0.83 |
| chr9_79719492_79719780 | 0.37 |
Col12a1 |
collagen, type XII, alpha 1 |
805 |
0.61 |
| chr19_55549970_55550134 | 0.37 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
169038 |
0.03 |
| chr18_38176274_38176683 | 0.37 |
Pcdh1 |
protocadherin 1 |
26685 |
0.12 |
| chr11_117833930_117834097 | 0.37 |
Afmid |
arylformamidase |
1740 |
0.17 |
| chr14_120378403_120378554 | 0.37 |
Mbnl2 |
muscleblind like splicing factor 2 |
763 |
0.7 |
| chr15_73060279_73060482 | 0.36 |
Trappc9 |
trafficking protein particle complex 9 |
665 |
0.74 |
| chr2_3737881_3738032 | 0.36 |
Gm13185 |
predicted gene 13185 |
17221 |
0.16 |
| chr9_78508116_78508485 | 0.36 |
Gm47430 |
predicted gene, 47430 |
12246 |
0.11 |
| chr19_29668488_29668647 | 0.36 |
Ermp1 |
endoplasmic reticulum metallopeptidase 1 |
20152 |
0.17 |
| chr8_90831859_90832023 | 0.36 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
3085 |
0.18 |
| chr3_130058674_130059202 | 0.36 |
Sec24b |
Sec24 related gene family, member B (S. cerevisiae) |
1969 |
0.28 |
| chr6_51635883_51636035 | 0.36 |
Gm22914 |
predicted gene, 22914 |
14327 |
0.23 |
| chr5_72220168_72220334 | 0.36 |
Atp10d |
ATPase, class V, type 10D |
16898 |
0.18 |
| chr1_128613454_128613628 | 0.35 |
Cxcr4 |
chemokine (C-X-C motif) receptor 4 |
21248 |
0.2 |
| chr17_84405686_84406042 | 0.35 |
Gm24722 |
predicted gene, 24722 |
14435 |
0.2 |
| chr5_134014798_134014973 | 0.35 |
1700030N18Rik |
RIKEN cDNA 1700030N18 gene |
76566 |
0.08 |
| chr15_97229103_97229537 | 0.35 |
Pced1b |
PC-esterase domain containing 1B |
17787 |
0.19 |
| chr5_125506718_125506869 | 0.35 |
Aacs |
acetoacetyl-CoA synthetase |
644 |
0.65 |
| chr7_137312803_137312954 | 0.35 |
Ebf3 |
early B cell factor 3 |
1038 |
0.54 |
| chr1_88123301_88123583 | 0.35 |
Gm15372 |
predicted gene 15372 |
3334 |
0.08 |
| chrX_166458575_166458751 | 0.35 |
Trappc2 |
trafficking protein particle complex 2 |
13360 |
0.13 |
| chr16_26566034_26566204 | 0.35 |
Il1rap |
interleukin 1 receptor accessory protein |
15585 |
0.24 |
| chr7_43453405_43453618 | 0.34 |
Etfb |
electron transferring flavoprotein, beta polypeptide |
621 |
0.43 |
| chr6_139852445_139852613 | 0.34 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
8881 |
0.22 |
| chr17_55976882_55977076 | 0.34 |
Shd |
src homology 2 domain-containing transforming protein D |
889 |
0.35 |
| chr6_108837450_108837611 | 0.34 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
1120 |
0.56 |
| chr13_37854787_37854958 | 0.34 |
Rreb1 |
ras responsive element binding protein 1 |
3062 |
0.27 |
| chr1_36874724_36874959 | 0.34 |
Gm37506 |
predicted gene, 37506 |
2796 |
0.22 |
| chr11_111975792_111975963 | 0.34 |
Gm11679 |
predicted gene 11679 |
67701 |
0.13 |
| chr16_87439512_87439663 | 0.34 |
Rwdd2b |
RWD domain containing 2B |
974 |
0.45 |
| chr10_89627085_89627236 | 0.34 |
Slc17a8 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
5907 |
0.2 |
| chr10_66914558_66914709 | 0.34 |
1110002J07Rik |
RIKEN cDNA 1110002J07 gene |
3106 |
0.21 |
| chr2_94147420_94147582 | 0.33 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
10392 |
0.17 |
| chr6_24046716_24047018 | 0.33 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
61704 |
0.12 |
| chr8_93166590_93166741 | 0.33 |
Ces1d |
carboxylesterase 1D |
3310 |
0.17 |
| chr5_76924147_76924300 | 0.33 |
2310040G07Rik |
RIKEN cDNA 2310040G07 gene |
4149 |
0.15 |
| chr19_57537517_57537807 | 0.33 |
Gm50285 |
predicted gene, 50285 |
6561 |
0.15 |
| chr6_116896579_116896756 | 0.33 |
Gm43926 |
predicted gene, 43926 |
56852 |
0.12 |
| chr9_74702140_74702734 | 0.33 |
Gm27233 |
predicted gene 27233 |
6825 |
0.25 |
| chr8_116504568_116504772 | 0.33 |
Dynlrb2 |
dynein light chain roadblock-type 2 |
304 |
0.92 |
| chr6_114787500_114787689 | 0.33 |
Gm44331 |
predicted gene, 44331 |
1796 |
0.33 |
| chr11_16698294_16698448 | 0.33 |
Gm25698 |
predicted gene, 25698 |
34340 |
0.14 |
| chr16_24517847_24518116 | 0.33 |
Morf4l1-ps1 |
mortality factor 4 like 1, pseudogene 1 |
11413 |
0.25 |
| chr8_48663447_48663609 | 0.33 |
Tenm3 |
teneurin transmembrane protein 3 |
11156 |
0.25 |
| chr12_8507138_8507301 | 0.33 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
3982 |
0.18 |
| chr17_79999311_79999483 | 0.33 |
Gm41625 |
predicted gene, 41625 |
17735 |
0.14 |
| chr8_121646337_121646786 | 0.32 |
Zcchc14 |
zinc finger, CCHC domain containing 14 |
6340 |
0.12 |
| chr1_88138022_88138189 | 0.32 |
Ugt1a6a |
UDP glucuronosyltransferase 1 family, polypeptide A6A |
273 |
0.75 |
| chr2_58790472_58790686 | 0.32 |
Upp2 |
uridine phosphorylase 2 |
25254 |
0.18 |
| chr5_21175987_21176254 | 0.32 |
Gsap |
gamma-secretase activating protein |
10135 |
0.15 |
| chr10_82083691_82083842 | 0.32 |
BC024063 |
cDNA sequence BC024063 |
413 |
0.75 |
| chr11_79758158_79758325 | 0.32 |
9130204K15Rik |
RIKEN cDNA 9130204K15 gene |
24646 |
0.13 |
| chr7_100624440_100624643 | 0.32 |
Gm14382 |
predicted gene 14382 |
23 |
0.96 |
| chr10_4781737_4781912 | 0.32 |
Esr1 |
estrogen receptor 1 (alpha) |
69173 |
0.13 |
| chr2_15065214_15065379 | 0.32 |
Arl5b |
ADP-ribosylation factor-like 5B |
2843 |
0.2 |
| chr18_46656328_46656479 | 0.32 |
Gm3734 |
predicted gene 3734 |
25596 |
0.13 |
| chr19_40178466_40178780 | 0.32 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
8663 |
0.16 |
| chr16_77466840_77467144 | 0.32 |
Gm37063 |
predicted gene, 37063 |
9942 |
0.12 |
| chr1_67191363_67191567 | 0.32 |
Gm15668 |
predicted gene 15668 |
57735 |
0.12 |
| chr17_29060889_29061047 | 0.32 |
Gm41556 |
predicted gene, 41556 |
2739 |
0.13 |
| chr18_9672403_9672580 | 0.32 |
Colec12 |
collectin sub-family member 12 |
35104 |
0.09 |
| chr10_69205907_69206104 | 0.31 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
2547 |
0.26 |
| chr15_52377925_52378095 | 0.31 |
Gm41322 |
predicted gene, 41322 |
67112 |
0.1 |
| chr1_67143038_67143247 | 0.31 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
20116 |
0.22 |
| chr11_119046302_119046504 | 0.31 |
Cbx8 |
chromobox 8 |
5434 |
0.17 |
| chr11_28684227_28684558 | 0.31 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
2828 |
0.27 |
| chr9_43823107_43823578 | 0.31 |
Nectin1 |
nectin cell adhesion molecule 1 |
2395 |
0.29 |
| chr2_156203206_156203400 | 0.31 |
Phf20 |
PHD finger protein 20 |
6547 |
0.14 |
| chr4_6225587_6225787 | 0.31 |
Ubxn2b |
UBX domain protein 2B |
34589 |
0.15 |
| chr3_157731356_157731520 | 0.31 |
Gm33466 |
predicted gene, 33466 |
5030 |
0.27 |
| chr14_21138478_21138696 | 0.31 |
Adk |
adenosine kinase |
62435 |
0.12 |
| chr19_37456339_37456518 | 0.30 |
Gm23026 |
predicted gene, 23026 |
7318 |
0.13 |
| chr16_97752199_97752361 | 0.30 |
Ripk4 |
receptor-interacting serine-threonine kinase 4 |
11507 |
0.2 |
| chr13_46735806_46736396 | 0.30 |
Nup153 |
nucleoporin 153 |
8161 |
0.17 |
| chr1_9981778_9981956 | 0.30 |
Gm15818 |
predicted gene 15818 |
520 |
0.62 |
| chr13_52819486_52819637 | 0.30 |
BB123696 |
expressed sequence BB123696 |
62356 |
0.13 |
| chr12_87895766_87895919 | 0.30 |
Gm2022 |
predicted pseudogene 2022 |
472 |
0.56 |
| chr19_44111938_44112122 | 0.30 |
Cwf19l1 |
CWF19-like 1, cell cycle control (S. pombe) |
968 |
0.39 |
| chr13_31506338_31507020 | 0.30 |
Foxq1 |
forkhead box Q1 |
49455 |
0.1 |
| chr2_158076982_158077133 | 0.30 |
2010009K17Rik |
RIKEN cDNA 2010009K17 gene |
14740 |
0.15 |
| chr4_155137056_155137330 | 0.30 |
Morn1 |
MORN repeat containing 1 |
26503 |
0.13 |
| chr6_39241464_39242040 | 0.30 |
Gm43479 |
predicted gene 43479 |
1638 |
0.32 |
| chr2_72887557_72887708 | 0.30 |
Sp3 |
trans-acting transcription factor 3 |
53452 |
0.1 |
| chr6_100848852_100849036 | 0.30 |
Ppp4r2 |
protein phosphatase 4, regulatory subunit 2 |
3541 |
0.26 |
| chr16_11410695_11410993 | 0.30 |
Snx29 |
sorting nexin 29 |
4187 |
0.27 |
| chr11_100951927_100952102 | 0.30 |
Stat3 |
signal transducer and activator of transcription 3 |
12474 |
0.13 |
| chr6_97251308_97251476 | 0.30 |
Lmod3 |
leiomodin 3 (fetal) |
1367 |
0.4 |
| chr7_130645820_130646001 | 0.30 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
19 |
0.98 |
| chr8_90829727_90829881 | 0.30 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
948 |
0.46 |
| chr13_51646798_51647087 | 0.30 |
Gm22806 |
predicted gene, 22806 |
1257 |
0.35 |
| chr13_69531568_69531734 | 0.30 |
Tent4a |
terminal nucleotidyltransferase 4A |
781 |
0.51 |
| chr9_23224794_23225147 | 0.29 |
Bmper |
BMP-binding endothelial regulator |
1684 |
0.55 |
| chr19_44403590_44403745 | 0.29 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
3023 |
0.2 |
| chr18_39301788_39301939 | 0.29 |
Gm15336 |
predicted gene 15336 |
827 |
0.63 |
| chr2_79958014_79958183 | 0.29 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
30307 |
0.24 |
| chr14_70220268_70220419 | 0.29 |
Sorbs3 |
sorbin and SH3 domain containing 3 |
8354 |
0.12 |
| chr7_90138739_90138890 | 0.29 |
Gm45223 |
predicted gene 45223 |
6322 |
0.12 |
| chr6_90515371_90515537 | 0.29 |
Gm44236 |
predicted gene, 44236 |
2152 |
0.21 |
| chr14_8251423_8251762 | 0.29 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
1930 |
0.34 |
| chr12_98745962_98746154 | 0.29 |
Zc3h14 |
zinc finger CCCH type containing 14 |
906 |
0.45 |
| chr10_8951017_8951174 | 0.29 |
Gm48728 |
predicted gene, 48728 |
854 |
0.63 |
| chr1_185370452_185370690 | 0.29 |
Eprs |
glutamyl-prolyl-tRNA synthetase |
2974 |
0.15 |
| chr6_141942226_141942737 | 0.29 |
Slco1a1 |
solute carrier organic anion transporter family, member 1a1 |
4329 |
0.26 |
| chr2_90554210_90554639 | 0.29 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
26223 |
0.18 |
| chr9_36794561_36794712 | 0.29 |
Ei24 |
etoposide induced 2.4 mRNA |
2384 |
0.2 |
| chr12_110268616_110268767 | 0.29 |
Dio3os |
deiodinase, iodothyronine type III, opposite strand |
9058 |
0.11 |
| chr8_105169758_105169909 | 0.29 |
Cbfb |
core binding factor beta |
841 |
0.4 |
| chr6_116068553_116068726 | 0.29 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
4517 |
0.2 |
| chr19_55099918_55100172 | 0.29 |
Gm31356 |
predicted gene, 31356 |
543 |
0.49 |
| chrX_103288811_103289364 | 0.28 |
Gm19200 |
predicted gene, 19200 |
117 |
0.9 |
| chr14_60387250_60387782 | 0.28 |
Amer2 |
APC membrane recruitment 2 |
9230 |
0.22 |
| chr9_74530860_74531022 | 0.28 |
Gm28622 |
predicted gene 28622 |
30437 |
0.19 |
| chr1_38128833_38129005 | 0.28 |
Rev1 |
REV1, DNA directed polymerase |
602 |
0.7 |
| chr12_41024854_41025032 | 0.28 |
Immp2l |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
614 |
0.72 |
| chr5_125388300_125388726 | 0.28 |
Ubc |
ubiquitin C |
673 |
0.42 |
| chr16_29991132_29991283 | 0.28 |
Gm1968 |
predicted gene 1968 |
12023 |
0.16 |
| chr16_22238973_22239124 | 0.28 |
Gm26980 |
predicted gene, 26980 |
2127 |
0.2 |
| chr3_62392302_62392453 | 0.28 |
Arhgef26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
27291 |
0.21 |
| chr6_85851621_85851788 | 0.28 |
Ptges3-ps |
prostaglandin E synthase 3, pseudogene |
7724 |
0.09 |
| chr1_159231811_159232536 | 0.28 |
Cop1 |
COP1, E3 ubiquitin ligase |
147 |
0.95 |
| chr1_175624328_175624488 | 0.28 |
Fh1 |
fumarate hydratase 1 |
1165 |
0.45 |
| chr4_100258335_100258573 | 0.28 |
Gm12706 |
predicted gene 12706 |
103605 |
0.07 |
| chr2_91161315_91161528 | 0.28 |
Madd |
MAP-kinase activating death domain |
1064 |
0.38 |
| chr15_36275360_36275511 | 0.28 |
Rnf19a |
ring finger protein 19A |
7663 |
0.12 |
| chr5_102670562_102670781 | 0.28 |
Arhgap24 |
Rho GTPase activating protein 24 |
54302 |
0.17 |
| chr17_65940216_65940367 | 0.28 |
Twsg1 |
twisted gastrulation BMP signaling modulator 1 |
10824 |
0.13 |
| chr4_150867947_150868379 | 0.28 |
Errfi1 |
ERBB receptor feedback inhibitor 1 |
13090 |
0.13 |
| chr4_3574138_3574411 | 0.28 |
Tmem68 |
transmembrane protein 68 |
253 |
0.75 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.7 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.3 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.4 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 0.3 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.4 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.2 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.3 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 | 0.2 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 1.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0097459 | iron ion import into cell(GO:0097459) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.3 | GO:0052696 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.1 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0044704 | single-organism reproductive behavior(GO:0044704) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.2 | GO:0051084 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.4 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.0 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.0 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.0 | GO:0071351 | cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.0 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 0.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |