Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
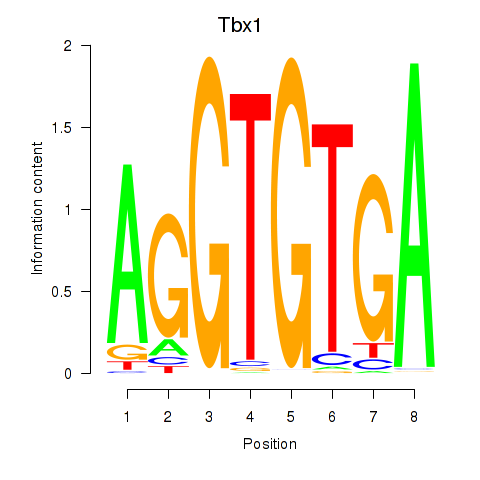
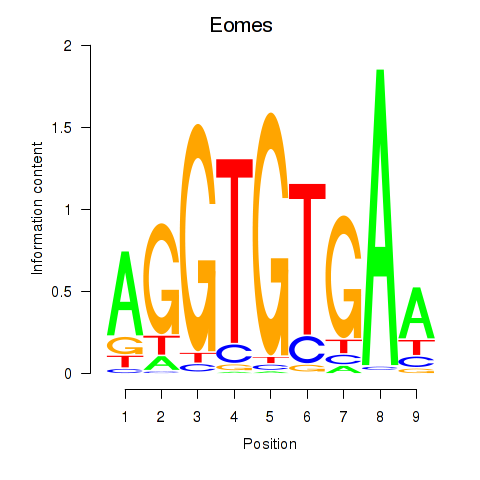
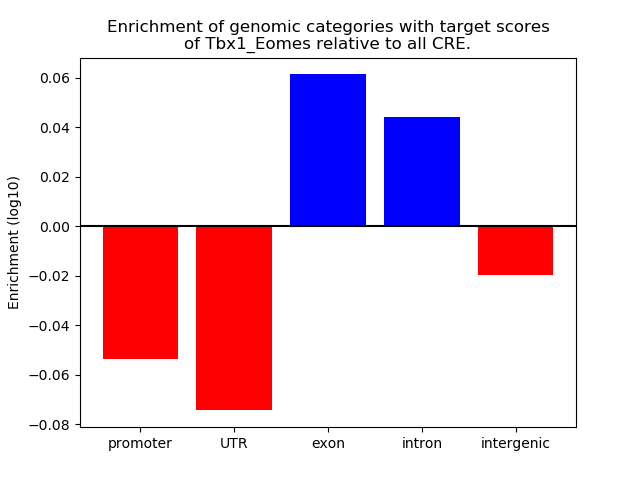
Results for Tbx1_Eomes
Z-value: 0.52
Transcription factors associated with Tbx1_Eomes
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx1
|
ENSMUSG00000009097.9 | T-box 1 |
|
Eomes
|
ENSMUSG00000032446.8 | eomesodermin |
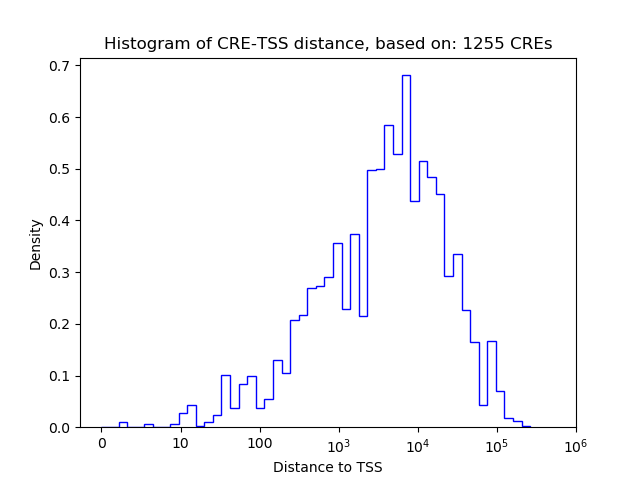
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_18590521_18590672 | Tbx1 | 75 | 0.941442 | 0.56 | 2.5e-01 | Click! |
| chr16_18574323_18574479 | Tbx1 | 8927 | 0.121537 | 0.24 | 6.5e-01 | Click! |
Activity of the Tbx1_Eomes motif across conditions
Conditions sorted by the z-value of the Tbx1_Eomes motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
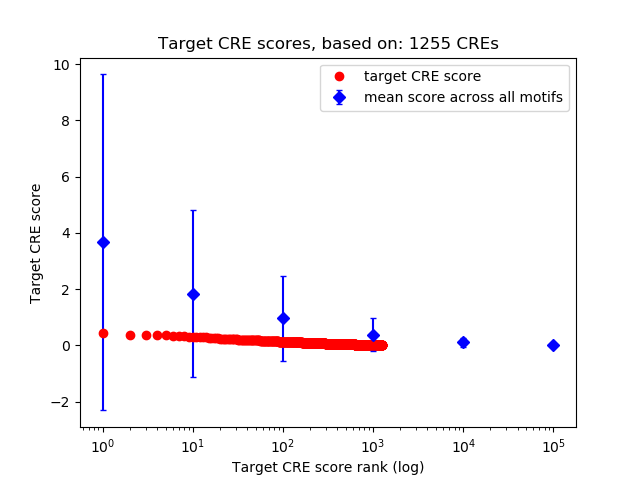
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_11861132_11861344 | 0.44 |
Gm25002 |
predicted gene, 25002 |
87649 |
0.07 |
| chr7_100658121_100658314 | 0.38 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
147 |
0.93 |
| chr1_171454993_171455144 | 0.37 |
F11r |
F11 receptor |
7782 |
0.1 |
| chr11_20826808_20826959 | 0.37 |
Lgalsl |
lectin, galactoside binding-like |
3872 |
0.18 |
| chr3_83996881_83997402 | 0.36 |
Tmem131l |
transmembrane 131 like |
28913 |
0.2 |
| chr1_193271420_193271628 | 0.35 |
G0s2 |
G0/G1 switch gene 2 |
1693 |
0.19 |
| chr1_84059698_84059849 | 0.34 |
Pid1 |
phosphotyrosine interaction domain containing 1 |
6520 |
0.27 |
| chr5_125296722_125296910 | 0.32 |
Scarb1 |
scavenger receptor class B, member 1 |
2707 |
0.22 |
| chr6_146473719_146473882 | 0.31 |
Gm15720 |
predicted gene 15720 |
10193 |
0.19 |
| chr5_75046920_75047087 | 0.30 |
Gm18593 |
predicted gene, 18593 |
1476 |
0.27 |
| chr2_104123492_104123669 | 0.30 |
A930018P22Rik |
RIKEN cDNA A930018P22 gene |
811 |
0.53 |
| chr9_110703228_110703412 | 0.30 |
Ccdc12 |
coiled-coil domain containing 12 |
6574 |
0.12 |
| chr11_86932862_86933027 | 0.29 |
Ypel2 |
yippee like 2 |
39080 |
0.14 |
| chr2_29437237_29437388 | 0.28 |
Gm24976 |
predicted gene, 24976 |
21882 |
0.15 |
| chr19_30146095_30146265 | 0.27 |
Gldc |
glycine decarboxylase |
949 |
0.57 |
| chr2_129101432_129101630 | 0.27 |
Polr1b |
polymerase (RNA) I polypeptide B |
504 |
0.68 |
| chr1_52388262_52388419 | 0.26 |
Gm23460 |
predicted gene, 23460 |
56204 |
0.11 |
| chr2_30386411_30386643 | 0.24 |
Miga2 |
mitoguardin 2 |
4583 |
0.1 |
| chr4_97013861_97014018 | 0.24 |
Gm27521 |
predicted gene, 27521 |
96919 |
0.08 |
| chr7_112498970_112499146 | 0.24 |
Parva |
parvin, alpha |
20643 |
0.2 |
| chr15_102181335_102181499 | 0.23 |
Csad |
cysteine sulfinic acid decarboxylase |
1556 |
0.24 |
| chr10_80972757_80972931 | 0.23 |
Gm3828 |
predicted gene 3828 |
18134 |
0.09 |
| chr11_86959315_86959659 | 0.23 |
Ypel2 |
yippee like 2 |
12537 |
0.18 |
| chr16_31233513_31233746 | 0.22 |
Gm41442 |
predicted gene, 41442 |
39 |
0.96 |
| chr19_42054767_42054919 | 0.21 |
4933411K16Rik |
RIKEN cDNA 4933411K16 gene |
2595 |
0.16 |
| chr3_31136392_31136619 | 0.21 |
Cldn11 |
claudin 11 |
13415 |
0.2 |
| chr6_112873464_112873750 | 0.21 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
44148 |
0.13 |
| chrX_73466714_73466865 | 0.21 |
Haus7 |
HAUS augmin-like complex, subunit 7 |
7745 |
0.12 |
| chr13_91765048_91765525 | 0.21 |
Gm27656 |
predicted gene, 27656 |
13180 |
0.14 |
| chr2_158199719_158199870 | 0.21 |
1700060C20Rik |
RIKEN cDNA 1700060C20 gene |
7786 |
0.15 |
| chr2_103454731_103454882 | 0.21 |
Elf5 |
E74-like factor 5 |
6068 |
0.19 |
| chr11_106581287_106581448 | 0.21 |
Tex2 |
testis expressed gene 2 |
767 |
0.66 |
| chr18_36726531_36726874 | 0.20 |
Cd14 |
CD14 antigen |
36 |
0.94 |
| chr17_56208632_56208824 | 0.20 |
Dpp9 |
dipeptidylpeptidase 9 |
480 |
0.64 |
| chr2_105901718_105901869 | 0.20 |
Elp4 |
elongator acetyltransferase complex subunit 4 |
2690 |
0.24 |
| chr12_82914756_82914931 | 0.20 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
24312 |
0.21 |
| chr6_28736579_28736738 | 0.19 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
28032 |
0.19 |
| chrX_103733822_103734171 | 0.19 |
Slc16a2 |
solute carrier family 16 (monocarboxylic acid transporters), member 2 |
193 |
0.92 |
| chr9_71028459_71028669 | 0.19 |
Lipc |
lipase, hepatic |
76338 |
0.09 |
| chr17_29093649_29093812 | 0.19 |
Cdkn1a |
cyclin-dependent kinase inhibitor 1A (P21) |
39 |
0.91 |
| chr11_50199089_50199272 | 0.19 |
Sqstm1 |
sequestosome 1 |
3774 |
0.13 |
| chr9_108297063_108297481 | 0.19 |
Amt |
aminomethyltransferase |
350 |
0.72 |
| chr2_167846760_167847264 | 0.18 |
Gm14319 |
predicted gene 14319 |
11573 |
0.16 |
| chr14_30906842_30907058 | 0.18 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
7179 |
0.11 |
| chr19_28928421_28928869 | 0.18 |
4430402I18Rik |
RIKEN cDNA 4430402I18 gene |
582 |
0.68 |
| chr9_115400503_115400904 | 0.18 |
Gm9487 |
predicted gene 9487 |
4446 |
0.15 |
| chr6_121859844_121860059 | 0.18 |
Mug1 |
murinoglobulin 1 |
18847 |
0.19 |
| chr1_58097131_58097333 | 0.18 |
Aox3 |
aldehyde oxidase 3 |
15898 |
0.18 |
| chr5_8983164_8983315 | 0.18 |
Crot |
carnitine O-octanoyltransferase |
1095 |
0.38 |
| chr1_166410038_166410239 | 0.18 |
Pogk |
pogo transposable element with KRAB domain |
275 |
0.88 |
| chr3_98311224_98311375 | 0.18 |
Phgdh |
3-phosphoglycerate dehydrogenase |
17314 |
0.14 |
| chr8_114555833_114556014 | 0.17 |
Gm16116 |
predicted gene 16116 |
38977 |
0.17 |
| chr6_58832026_58832202 | 0.17 |
Herc3 |
hect domain and RLD 3 |
253 |
0.93 |
| chr10_71183397_71183774 | 0.17 |
C730027H18Rik |
RIKEN cDNA C730027H18 gene |
1533 |
0.26 |
| chr2_126569048_126569199 | 0.17 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
4363 |
0.22 |
| chr3_28025480_28025631 | 0.17 |
Pld1 |
phospholipase D1 |
5707 |
0.3 |
| chr11_86976287_86976438 | 0.17 |
Ypel2 |
yippee like 2 |
4338 |
0.2 |
| chr6_5512042_5512193 | 0.17 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
15808 |
0.27 |
| chr14_63265088_63265239 | 0.17 |
Gata4 |
GATA binding protein 4 |
5961 |
0.19 |
| chr8_88863392_88863746 | 0.17 |
Gm45504 |
predicted gene 45504 |
495 |
0.85 |
| chr6_87846724_87847004 | 0.16 |
Cnbp |
cellular nucleic acid binding protein |
547 |
0.54 |
| chr15_75996647_75996807 | 0.16 |
Mapk15 |
mitogen-activated protein kinase 15 |
14 |
0.94 |
| chr11_101372782_101373038 | 0.16 |
G6pc |
glucose-6-phosphatase, catalytic |
5349 |
0.06 |
| chr8_120495609_120495777 | 0.16 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
7246 |
0.15 |
| chr4_144896227_144896412 | 0.16 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
3100 |
0.28 |
| chr2_26405907_26406058 | 0.15 |
Inpp5e |
inositol polyphosphate-5-phosphatase E |
3145 |
0.11 |
| chr2_163103270_163103612 | 0.15 |
Gm22174 |
predicted gene, 22174 |
10542 |
0.12 |
| chr19_3329840_3329991 | 0.15 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
3810 |
0.15 |
| chr1_182500736_182501185 | 0.15 |
Gm37069 |
predicted gene, 37069 |
6804 |
0.14 |
| chr10_21448325_21448476 | 0.15 |
Gm48386 |
predicted gene, 48386 |
2764 |
0.21 |
| chr16_46463130_46463327 | 0.15 |
Nectin3 |
nectin cell adhesion molecule 3 |
14718 |
0.27 |
| chr2_128567912_128568063 | 0.15 |
Gm14006 |
predicted gene 14006 |
9753 |
0.13 |
| chr10_61143598_61144049 | 0.15 |
Sgpl1 |
sphingosine phosphate lyase 1 |
3075 |
0.2 |
| chr1_155888091_155888242 | 0.15 |
Cep350 |
centrosomal protein 350 |
2152 |
0.22 |
| chr2_92056065_92056235 | 0.15 |
2900072N19Rik |
RIKEN cDNA 2900072N19 gene |
3219 |
0.2 |
| chr11_109488275_109488451 | 0.15 |
Arsg |
arylsulfatase G |
2757 |
0.18 |
| chr5_135139962_135140114 | 0.15 |
Mlxipl |
MLX interacting protein-like |
6407 |
0.11 |
| chr5_121732457_121732668 | 0.15 |
Gm16552 |
predicted gene 16552 |
5159 |
0.13 |
| chr6_94182054_94182207 | 0.15 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
100895 |
0.08 |
| chr8_124589098_124589249 | 0.14 |
Capn9 |
calpain 9 |
13060 |
0.16 |
| chr11_79670395_79670546 | 0.14 |
Rab11fip4os2 |
RAB11 family interacting protein 4 (class II), opposite strand 2 |
4613 |
0.13 |
| chr11_53420114_53420567 | 0.14 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
2830 |
0.12 |
| chr4_107159322_107159488 | 0.14 |
Tceanc2 |
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
18263 |
0.12 |
| chr17_31383992_31384162 | 0.14 |
Pde9a |
phosphodiesterase 9A |
2133 |
0.25 |
| chr14_65832955_65833117 | 0.14 |
Esco2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
872 |
0.57 |
| chr2_57332734_57332885 | 0.14 |
Gm13535 |
predicted gene 13535 |
57123 |
0.11 |
| chr13_34645504_34645908 | 0.14 |
Pxdc1 |
PX domain containing 1 |
2862 |
0.18 |
| chr3_31136684_31136995 | 0.14 |
Cldn11 |
claudin 11 |
13081 |
0.2 |
| chr5_145993052_145993431 | 0.14 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
1598 |
0.28 |
| chr10_61337817_61337968 | 0.14 |
Pald1 |
phosphatase domain containing, paladin 1 |
4435 |
0.15 |
| chr11_79230285_79230459 | 0.13 |
Wsb1 |
WD repeat and SOCS box-containing 1 |
12577 |
0.16 |
| chr17_86497669_86497820 | 0.13 |
Prkce |
protein kinase C, epsilon |
4417 |
0.26 |
| chr5_53437072_53437250 | 0.13 |
Gm26486 |
predicted gene, 26486 |
18588 |
0.17 |
| chr15_102442382_102442533 | 0.13 |
Amhr2 |
anti-Mullerian hormone type 2 receptor |
2910 |
0.13 |
| chr13_104706600_104706768 | 0.13 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
21753 |
0.23 |
| chr4_134060274_134060433 | 0.13 |
Crybg2 |
crystallin beta-gamma domain containing 2 |
462 |
0.7 |
| chr3_98297519_98297734 | 0.13 |
Gm43189 |
predicted gene 43189 |
10638 |
0.14 |
| chr11_85530391_85530542 | 0.13 |
Bcas3 |
breast carcinoma amplified sequence 3 |
20828 |
0.21 |
| chr7_141070384_141070555 | 0.13 |
B4galnt4 |
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
133 |
0.9 |
| chr19_30202160_30202660 | 0.13 |
Gldc |
glycine decarboxylase |
26981 |
0.15 |
| chr19_23066702_23066853 | 0.13 |
Gm50136 |
predicted gene, 50136 |
5304 |
0.21 |
| chr1_191822042_191822193 | 0.13 |
Nek2 |
NIMA (never in mitosis gene a)-related expressed kinase 2 |
65 |
0.58 |
| chr5_137986079_137986259 | 0.13 |
Azgp1 |
alpha-2-glycoprotein 1, zinc |
4601 |
0.1 |
| chr15_97161836_97161987 | 0.13 |
Gm32885 |
predicted gene, 32885 |
5311 |
0.27 |
| chr9_77749808_77749964 | 0.13 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
4649 |
0.16 |
| chr13_94708250_94708413 | 0.13 |
Gm32351 |
predicted gene, 32351 |
5613 |
0.17 |
| chr6_115464997_115465148 | 0.13 |
Gm44079 |
predicted gene, 44079 |
82 |
0.97 |
| chr19_29125266_29125417 | 0.12 |
Mir101b |
microRNA 101b |
9938 |
0.14 |
| chr5_117275916_117276216 | 0.12 |
Pebp1 |
phosphatidylethanolamine binding protein 1 |
11459 |
0.11 |
| chr14_73045910_73046229 | 0.12 |
Cysltr2 |
cysteinyl leukotriene receptor 2 |
3003 |
0.3 |
| chr10_30810298_30810917 | 0.12 |
Ncoa7 |
nuclear receptor coactivator 7 |
7281 |
0.16 |
| chr9_110390754_110390978 | 0.12 |
Ptpn23 |
protein tyrosine phosphatase, non-receptor type 23 |
391 |
0.77 |
| chr6_122216632_122216783 | 0.12 |
Mug-ps1 |
murinoglobulin, pseudogene 1 |
12353 |
0.15 |
| chr1_167374753_167374904 | 0.12 |
Aldh9a1 |
aldehyde dehydrogenase 9, subfamily A1 |
10662 |
0.13 |
| chr5_72654251_72654431 | 0.12 |
Nipal1 |
NIPA-like domain containing 1 |
6499 |
0.12 |
| chr9_24378970_24379135 | 0.12 |
Gm22380 |
predicted gene, 22380 |
27934 |
0.15 |
| chr5_122494527_122494678 | 0.12 |
Gm43360 |
predicted gene 43360 |
306 |
0.77 |
| chr1_178314559_178314724 | 0.12 |
B230369F24Rik |
RIKEN cDNA B230369F24 gene |
3908 |
0.13 |
| chr10_28178649_28178847 | 0.12 |
Gm22370 |
predicted gene, 22370 |
35373 |
0.21 |
| chr16_16888784_16888935 | 0.12 |
Top3b |
topoisomerase (DNA) III beta |
1020 |
0.34 |
| chr15_89177223_89177386 | 0.12 |
Plxnb2 |
plexin B2 |
3484 |
0.11 |
| chrX_143067335_143067542 | 0.12 |
Rtl9 |
retrotransposon Gag like 9 |
32156 |
0.2 |
| chr12_79442844_79442995 | 0.12 |
Rad51b |
RAD51 paralog B |
115566 |
0.06 |
| chr5_145743023_145743196 | 0.11 |
Gm43626 |
predicted gene 43626 |
17436 |
0.15 |
| chr11_120526901_120527125 | 0.11 |
Gm11789 |
predicted gene 11789 |
3541 |
0.08 |
| chr1_73909397_73909605 | 0.11 |
Tns1 |
tensin 1 |
18716 |
0.18 |
| chr11_75437280_75437567 | 0.11 |
Serpinf2 |
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
1477 |
0.2 |
| chr1_52816740_52816904 | 0.11 |
Inpp1 |
inositol polyphosphate-1-phosphatase |
629 |
0.71 |
| chr9_43756806_43756967 | 0.11 |
Gm30015 |
predicted gene, 30015 |
10786 |
0.15 |
| chr11_7193213_7193425 | 0.11 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
4463 |
0.19 |
| chr5_63910906_63911057 | 0.11 |
Gm15819 |
predicted gene 15819 |
12920 |
0.16 |
| chr16_24424101_24424681 | 0.11 |
Gm24440 |
predicted gene, 24440 |
17974 |
0.17 |
| chr11_22843395_22843546 | 0.11 |
Gm23772 |
predicted gene, 23772 |
3398 |
0.15 |
| chr19_37860664_37860851 | 0.11 |
Gm4757 |
predicted gene 4757 |
9513 |
0.21 |
| chr1_181005812_181005979 | 0.11 |
Ephx1 |
epoxide hydrolase 1, microsomal |
9725 |
0.09 |
| chr18_44633494_44633779 | 0.11 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
28029 |
0.19 |
| chr15_100414144_100414311 | 0.11 |
Gm5475 |
predicted gene 5475 |
7920 |
0.11 |
| chr2_10674812_10674992 | 0.11 |
Mir669h |
microRNA 669h |
156747 |
0.01 |
| chr11_103011230_103011391 | 0.11 |
Dcakd |
dephospho-CoA kinase domain containing |
5858 |
0.13 |
| chr17_46484376_46484539 | 0.11 |
Ttbk1 |
tau tubulin kinase 1 |
162 |
0.91 |
| chr8_122174579_122174745 | 0.11 |
Zfp469 |
zinc finger protein 469 |
83958 |
0.07 |
| chr19_20645058_20645209 | 0.11 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
43172 |
0.15 |
| chr5_106313971_106314130 | 0.11 |
Gm32921 |
predicted gene, 32921 |
7290 |
0.16 |
| chr5_117368668_117368892 | 0.11 |
Wsb2 |
WD repeat and SOCS box-containing 2 |
5258 |
0.12 |
| chr4_126067500_126067685 | 0.11 |
Rpl28-ps3 |
ribosomal protein L28, pseudogene 3 |
2667 |
0.17 |
| chr4_109143752_109143931 | 0.11 |
Osbpl9 |
oxysterol binding protein-like 9 |
12769 |
0.21 |
| chr17_35725713_35725941 | 0.10 |
Gm20443 |
predicted gene 20443 |
13979 |
0.08 |
| chr3_145759243_145759445 | 0.10 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
669 |
0.72 |
| chr11_70741343_70741494 | 0.10 |
Gm12320 |
predicted gene 12320 |
13733 |
0.08 |
| chr6_143635075_143635226 | 0.10 |
1700060C16Rik |
RIKEN cDNA 1700060C16 gene |
16138 |
0.27 |
| chr7_3221770_3221921 | 0.10 |
Mir295 |
microRNA 295 |
1072 |
0.16 |
| chr15_88870480_88870800 | 0.10 |
Pim3 |
proviral integration site 3 |
5720 |
0.14 |
| chr5_45513018_45513181 | 0.10 |
Lap3 |
leucine aminopeptidase 3 |
3890 |
0.14 |
| chr2_167755014_167755358 | 0.10 |
Gm14321 |
predicted gene 14321 |
22281 |
0.12 |
| chr5_72652767_72652918 | 0.10 |
Nipal1 |
NIPA-like domain containing 1 |
5000 |
0.13 |
| chr10_81560885_81561091 | 0.10 |
Tle5 |
TLE family member 5, transcriptional modulator |
56 |
0.93 |
| chr9_31302317_31302496 | 0.10 |
Prdm10 |
PR domain containing 10 |
11147 |
0.16 |
| chr7_100120840_100121149 | 0.10 |
Pold3 |
polymerase (DNA-directed), delta 3, accessory subunit |
84 |
0.96 |
| chr19_43893023_43893176 | 0.10 |
Dnmbp |
dynamin binding protein |
2408 |
0.22 |
| chr6_8510307_8510729 | 0.10 |
A430035B10Rik |
RIKEN cDNA A430035B10 gene |
88 |
0.91 |
| chr14_47475021_47475196 | 0.10 |
Fbxo34 |
F-box protein 34 |
931 |
0.45 |
| chr19_3624054_3624227 | 0.10 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
8348 |
0.16 |
| chr10_127160387_127160730 | 0.10 |
B4galnt1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
4667 |
0.09 |
| chr8_33896084_33896490 | 0.10 |
Rbpms |
RNA binding protein gene with multiple splicing |
4523 |
0.2 |
| chr19_3687091_3687242 | 0.10 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
602 |
0.61 |
| chr2_72866286_72866437 | 0.10 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
52644 |
0.12 |
| chr2_30363854_30364005 | 0.10 |
Miga2 |
mitoguardin 2 |
304 |
0.79 |
| chr11_119913357_119913544 | 0.10 |
Chmp6 |
charged multivesicular body protein 6 |
9 |
0.97 |
| chr11_102211338_102211489 | 0.10 |
Hdac5 |
histone deacetylase 5 |
7515 |
0.09 |
| chr11_100859548_100859739 | 0.10 |
Stat5a |
signal transducer and activator of transcription 5A |
13 |
0.97 |
| chr18_6208229_6208380 | 0.10 |
Gm17036 |
predicted gene 17036 |
5609 |
0.19 |
| chr16_35055198_35055367 | 0.09 |
Hacd2 |
3-hydroxyacyl-CoA dehydratase 2 |
6667 |
0.21 |
| chr6_56919800_56919965 | 0.09 |
Nt5c3 |
5'-nucleotidase, cytosolic III |
3842 |
0.15 |
| chr2_27471316_27471526 | 0.09 |
Brd3 |
bromodomain containing 3 |
3760 |
0.17 |
| chr1_134344955_134345133 | 0.09 |
Tmem183a |
transmembrane protein 183A |
5261 |
0.14 |
| chr7_84135967_84136134 | 0.09 |
Abhd17c |
abhydrolase domain containing 17C |
11765 |
0.15 |
| chr1_74304286_74304581 | 0.09 |
Tmbim1 |
transmembrane BAX inhibitor motif containing 1 |
28 |
0.94 |
| chr8_103538740_103538926 | 0.09 |
Gm45277 |
predicted gene 45277 |
24790 |
0.23 |
| chr8_124291752_124291934 | 0.09 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
3260 |
0.21 |
| chr4_155207879_155208052 | 0.09 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
14570 |
0.17 |
| chr4_107159764_107160176 | 0.09 |
Tceanc2 |
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
17698 |
0.12 |
| chr6_30302088_30302239 | 0.09 |
Ube2h |
ubiquitin-conjugating enzyme E2H |
2359 |
0.22 |
| chr8_126727960_126728150 | 0.09 |
Gm45805 |
predicted gene 45805 |
30279 |
0.2 |
| chr6_137747661_137747817 | 0.09 |
Strap |
serine/threonine kinase receptor associated protein |
1486 |
0.45 |
| chr5_89416345_89416742 | 0.09 |
Gc |
vitamin D binding protein |
19085 |
0.24 |
| chr17_84990758_84990958 | 0.09 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
2815 |
0.25 |
| chr11_101554799_101555209 | 0.09 |
Nbr1 |
NBR1, autophagy cargo receptor |
862 |
0.39 |
| chr3_54723479_54723643 | 0.09 |
Exosc8 |
exosome component 8 |
5897 |
0.14 |
| chr15_91276343_91276512 | 0.09 |
CN725425 |
cDNA sequence CN725425 |
44849 |
0.16 |
| chr7_112187666_112187837 | 0.09 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
28694 |
0.2 |
| chr13_64361503_64361746 | 0.09 |
Gm49230 |
predicted gene, 49230 |
113 |
0.86 |
| chr15_34510659_34511041 | 0.09 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
943 |
0.43 |
| chr2_132240536_132240711 | 0.09 |
Tmem230 |
transmembrane protein 230 |
7031 |
0.15 |
| chr4_137875186_137875337 | 0.08 |
Gm13012 |
predicted gene 13012 |
8151 |
0.2 |
| chr2_6074858_6075460 | 0.08 |
Upf2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
24732 |
0.16 |
| chr12_80832436_80832620 | 0.08 |
Susd6 |
sushi domain containing 6 |
36180 |
0.11 |
| chrX_163908542_163908757 | 0.08 |
Ap1s2 |
adaptor-related protein complex 1, sigma 2 subunit |
368 |
0.89 |
| chr8_72222949_72223319 | 0.08 |
Fam32a |
family with sequence similarity 32, member A |
3370 |
0.12 |
| chr11_109500636_109500796 | 0.08 |
Gm22378 |
predicted gene, 22378 |
892 |
0.5 |
| chrX_60339140_60339308 | 0.08 |
Mir505 |
microRNA 505 |
55268 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.2 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |