Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
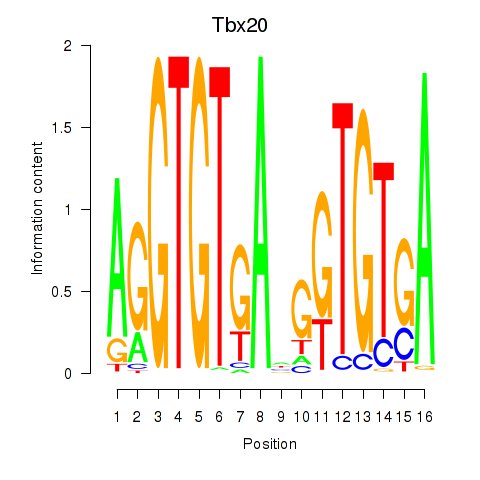
Results for Tbx20
Z-value: 0.88
Transcription factors associated with Tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx20
|
ENSMUSG00000031965.8 | T-box 20 |
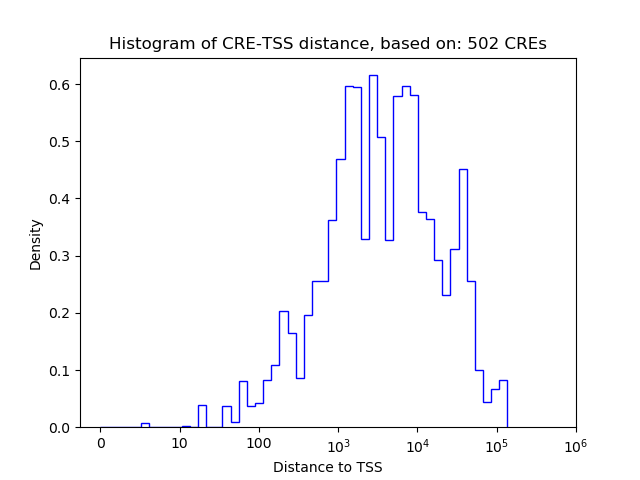
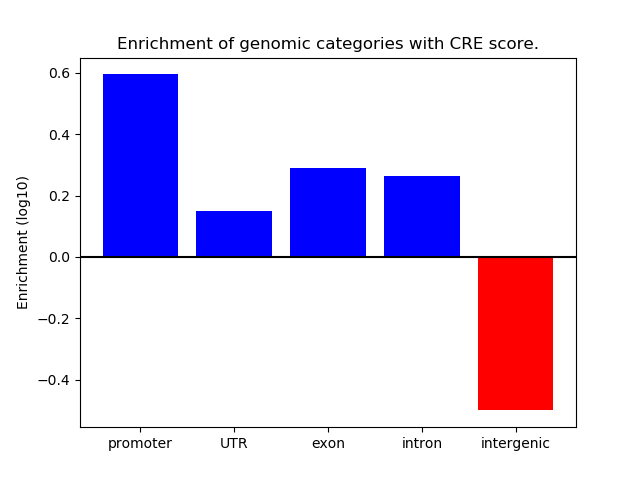
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_24774302_24774460 | Tbx20 | 78 | 0.966420 | 0.32 | 5.4e-01 | Click! |
Activity of the Tbx20 motif across conditions
Conditions sorted by the z-value of the Tbx20 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
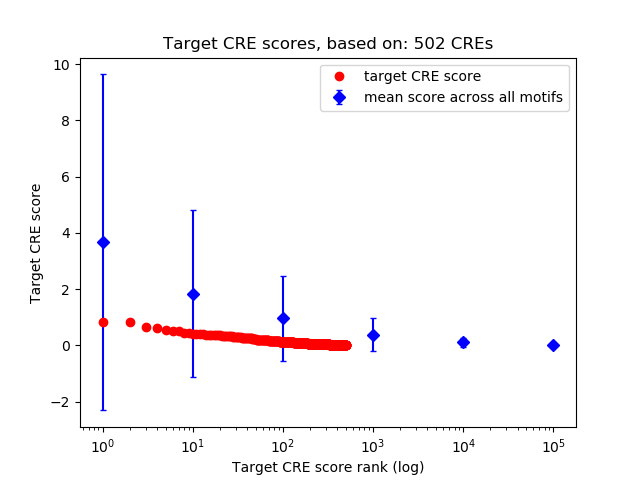
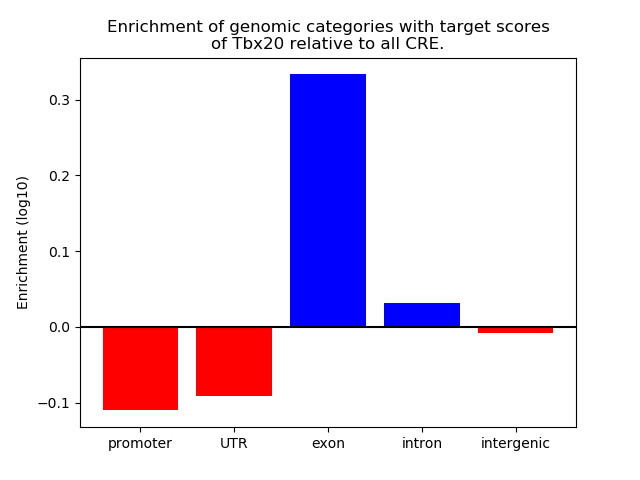
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_55468108_55468290 | 0.84 |
G3bp1 |
GTPase activating protein (SH3 domain) binding protein 1 |
1486 |
0.25 |
| chr15_73731010_73731168 | 0.82 |
Gm22106 |
predicted gene, 22106 |
1925 |
0.28 |
| chr5_43865480_43865631 | 0.67 |
Cd38 |
CD38 antigen |
2998 |
0.14 |
| chr3_146616407_146616685 | 0.60 |
Dnase2b |
deoxyribonuclease II beta |
950 |
0.46 |
| chr13_44232139_44232290 | 0.54 |
Gm47781 |
predicted gene, 47781 |
184 |
0.94 |
| chr15_3521825_3522033 | 0.52 |
Ghr |
growth hormone receptor |
50285 |
0.16 |
| chr9_66624894_66625176 | 0.49 |
Usp3 |
ubiquitin specific peptidase 3 |
31893 |
0.16 |
| chr2_132639230_132639433 | 0.45 |
AU019990 |
expressed sequence AU019990 |
6550 |
0.14 |
| chrX_93584463_93584645 | 0.44 |
Gm23557 |
predicted gene, 23557 |
10686 |
0.19 |
| chr9_83251916_83252091 | 0.42 |
Gm27216 |
predicted gene 27216 |
2019 |
0.35 |
| chr12_73408646_73408959 | 0.41 |
D830013O20Rik |
RIKEN cDNA D830013O20 gene |
755 |
0.63 |
| chr4_118437515_118437694 | 0.40 |
Cdc20 |
cell division cycle 20 |
252 |
0.84 |
| chr4_133555852_133556164 | 0.39 |
Nr0b2 |
nuclear receptor subfamily 0, group B, member 2 |
2632 |
0.14 |
| chr8_103180795_103180954 | 0.38 |
Gm45391 |
predicted gene 45391 |
35928 |
0.21 |
| chr1_156978787_156978968 | 0.38 |
4930439D14Rik |
RIKEN cDNA 4930439D14 gene |
39039 |
0.12 |
| chr8_22505624_22505807 | 0.38 |
Slc20a2 |
solute carrier family 20, member 2 |
410 |
0.8 |
| chr5_145993052_145993431 | 0.37 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
1598 |
0.28 |
| chr7_120177637_120177817 | 0.36 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
3869 |
0.16 |
| chr3_89142152_89142303 | 0.35 |
Pklr |
pyruvate kinase liver and red blood cell |
5604 |
0.07 |
| chr1_93067493_93067644 | 0.35 |
Kif1a |
kinesin family member 1A |
1190 |
0.41 |
| chr8_119445523_119446899 | 0.35 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
508 |
0.74 |
| chr13_39657075_39657270 | 0.34 |
Gm47352 |
predicted gene, 47352 |
7661 |
0.21 |
| chr18_60627313_60627464 | 0.33 |
Synpo |
synaptopodin |
2465 |
0.26 |
| chr19_48656780_48656950 | 0.32 |
Sorcs3 |
sortilin-related VPS10 domain containing receptor 3 |
47351 |
0.19 |
| chr1_9906956_9907361 | 0.32 |
Mcmdc2 |
minichromosome maintenance domain containing 2 |
1480 |
0.29 |
| chr13_29049878_29050059 | 0.32 |
A330102I10Rik |
RIKEN cDNA A330102I10 gene |
33260 |
0.19 |
| chr13_80896624_80897028 | 0.31 |
Arrdc3 |
arrestin domain containing 3 |
6308 |
0.17 |
| chr2_70816094_70816245 | 0.31 |
Tlk1 |
tousled-like kinase 1 |
9059 |
0.21 |
| chr6_17178805_17178999 | 0.30 |
Gm4876 |
predicted gene 4876 |
7069 |
0.21 |
| chr17_5387030_5387190 | 0.29 |
Gm29050 |
predicted gene 29050 |
1653 |
0.37 |
| chr11_90006065_90006424 | 0.29 |
Pctp |
phosphatidylcholine transfer protein |
3350 |
0.29 |
| chr2_163564305_163564477 | 0.29 |
Hnf4a |
hepatic nuclear factor 4, alpha |
14308 |
0.12 |
| chr19_3329840_3329991 | 0.28 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
3810 |
0.15 |
| chr10_87536757_87536908 | 0.28 |
Pah |
phenylalanine hydroxylase |
9841 |
0.21 |
| chr2_122298688_122298852 | 0.28 |
Duoxa2 |
dual oxidase maturation factor 2 |
130 |
0.72 |
| chr15_82689760_82690423 | 0.28 |
Cyp2d37-ps |
cytochrome P450, family 2, subfamily d, polypeptide 37, pseudogene |
2910 |
0.12 |
| chr9_60837448_60837607 | 0.27 |
Gm9869 |
predicted gene 9869 |
672 |
0.71 |
| chrX_102504843_102504994 | 0.27 |
Hdac8 |
histone deacetylase 8 |
163 |
0.96 |
| chr11_16896137_16896457 | 0.27 |
Egfr |
epidermal growth factor receptor |
8888 |
0.2 |
| chr3_10315469_10315691 | 0.27 |
Impa1 |
inositol (myo)-1(or 4)-monophosphatase 1 |
1072 |
0.33 |
| chr2_90496134_90496285 | 0.26 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
16959 |
0.17 |
| chr18_35681549_35681863 | 0.26 |
Dnajc18 |
DnaJ heat shock protein family (Hsp40) member C18 |
5037 |
0.1 |
| chr9_63184988_63185331 | 0.26 |
Skor1 |
SKI family transcriptional corepressor 1 |
36198 |
0.15 |
| chr11_16906555_16906817 | 0.25 |
Egfr |
epidermal growth factor receptor |
1501 |
0.41 |
| chr2_38531241_38531405 | 0.25 |
Gm35808 |
predicted gene, 35808 |
4384 |
0.14 |
| chr5_76044220_76044385 | 0.23 |
Gm32780 |
predicted gene, 32780 |
1204 |
0.44 |
| chr11_16879987_16880147 | 0.22 |
Egfr |
epidermal growth factor receptor |
1917 |
0.36 |
| chr1_52388262_52388419 | 0.22 |
Gm23460 |
predicted gene, 23460 |
56204 |
0.11 |
| chr16_36083499_36083674 | 0.21 |
Ccdc58 |
coiled-coil domain containing 58 |
2311 |
0.17 |
| chr7_116205581_116205767 | 0.21 |
Plekha7 |
pleckstrin homology domain containing, family A member 7 |
7113 |
0.22 |
| chr10_125327017_125327436 | 0.20 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
1309 |
0.51 |
| chr5_110057791_110057958 | 0.20 |
Gm17655 |
predicted gene, 17655 |
11388 |
0.11 |
| chr19_37262158_37262416 | 0.19 |
Ide |
insulin degrading enzyme |
28591 |
0.11 |
| chr8_69994802_69994953 | 0.19 |
Gatad2a |
GATA zinc finger domain containing 2A |
1356 |
0.31 |
| chr8_117692083_117692234 | 0.19 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
9746 |
0.13 |
| chr5_65624489_65624645 | 0.19 |
Gm42729 |
predicted gene 42729 |
633 |
0.52 |
| chr13_24949534_24949740 | 0.19 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
6217 |
0.14 |
| chr19_47121147_47121319 | 0.19 |
Pdcd11 |
programmed cell death 11 |
4917 |
0.11 |
| chr11_99054515_99054937 | 0.19 |
Igfbp4 |
insulin-like growth factor binding protein 4 |
7415 |
0.13 |
| chr5_72652767_72652918 | 0.19 |
Nipal1 |
NIPA-like domain containing 1 |
5000 |
0.13 |
| chr17_13684368_13684541 | 0.18 |
Gm16046 |
predicted gene 16046 |
938 |
0.5 |
| chr2_103476872_103477023 | 0.18 |
Cat |
catalase |
8178 |
0.19 |
| chr3_103904824_103904993 | 0.18 |
Gm15602 |
predicted gene 15602 |
1945 |
0.18 |
| chr6_108668179_108668420 | 0.18 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
5253 |
0.18 |
| chr13_56630439_56630647 | 0.18 |
Tgfbi |
transforming growth factor, beta induced |
670 |
0.72 |
| chr19_10596904_10597210 | 0.18 |
Tkfc |
triokinase, FMN cyclase |
7201 |
0.09 |
| chr6_22026861_22027316 | 0.18 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
1145 |
0.61 |
| chr14_121315001_121315152 | 0.18 |
Stk24 |
serine/threonine kinase 24 |
11601 |
0.21 |
| chr7_79659882_79660033 | 0.17 |
Ticrr |
TOPBP1-interacting checkpoint and replication regulator |
239 |
0.88 |
| chr14_75337969_75338142 | 0.17 |
Zc3h13 |
zinc finger CCCH type containing 13 |
17 |
0.98 |
| chr4_155316923_155317235 | 0.17 |
Prkcz |
protein kinase C, zeta |
15908 |
0.15 |
| chr1_187662199_187662362 | 0.17 |
Gm37929 |
predicted gene, 37929 |
1081 |
0.53 |
| chr2_164425523_164425863 | 0.17 |
Sdc4 |
syndecan 4 |
17493 |
0.08 |
| chr3_108112650_108112909 | 0.17 |
Gnat2 |
guanine nucleotide binding protein, alpha transducing 2 |
15536 |
0.08 |
| chr1_133328332_133328498 | 0.17 |
Kiss1 |
KiSS-1 metastasis-suppressor |
1203 |
0.34 |
| chr14_54896568_54896719 | 0.16 |
Pabpn1 |
poly(A) binding protein, nuclear 1 |
1810 |
0.14 |
| chr7_19824719_19824878 | 0.16 |
Bcl3 |
B cell leukemia/lymphoma 3 |
2028 |
0.13 |
| chr7_115624565_115624753 | 0.16 |
Sox6 |
SRY (sex determining region Y)-box 6 |
26763 |
0.26 |
| chr1_171501634_171501795 | 0.16 |
Gm24871 |
predicted gene, 24871 |
1449 |
0.19 |
| chr12_84969491_84969654 | 0.15 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
1252 |
0.29 |
| chr12_110757292_110757448 | 0.15 |
Wdr20 |
WD repeat domain 20 |
19161 |
0.13 |
| chr14_47983657_47983808 | 0.15 |
Gm49304 |
predicted gene, 49304 |
10071 |
0.15 |
| chr13_69701514_69701677 | 0.15 |
Gm48819 |
predicted gene, 48819 |
3784 |
0.13 |
| chr8_10854076_10854227 | 0.15 |
Gm32540 |
predicted gene, 32540 |
12035 |
0.12 |
| chr4_129575301_129575464 | 0.15 |
Lck |
lymphocyte protein tyrosine kinase |
1741 |
0.16 |
| chr11_86200523_86200674 | 0.14 |
Brip1 |
BRCA1 interacting protein C-terminal helicase 1 |
566 |
0.53 |
| chr6_125482442_125482599 | 0.14 |
Gm28809 |
predicted gene 28809 |
576 |
0.69 |
| chr8_117085576_117085921 | 0.14 |
Pkd1l2 |
polycystic kidney disease 1 like 2 |
3299 |
0.21 |
| chr5_151195640_151195791 | 0.14 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
5446 |
0.25 |
| chr8_114154739_114154891 | 0.14 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
21173 |
0.24 |
| chr11_53818731_53818912 | 0.14 |
Gm12216 |
predicted gene 12216 |
22946 |
0.11 |
| chr5_3420296_3420447 | 0.14 |
Cdk6 |
cyclin-dependent kinase 6 |
50578 |
0.1 |
| chr2_126554079_126554230 | 0.13 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
564 |
0.77 |
| chr4_106688753_106689064 | 0.13 |
Ttc4 |
tetratricopeptide repeat domain 4 |
9964 |
0.13 |
| chr14_76657488_76657670 | 0.13 |
1700108F19Rik |
RIKEN cDNA 1700108F19 gene |
15384 |
0.19 |
| chr7_110824083_110824234 | 0.13 |
Rnf141 |
ring finger protein 141 |
2550 |
0.22 |
| chr4_105171470_105171650 | 0.13 |
Plpp3 |
phospholipid phosphatase 3 |
14213 |
0.26 |
| chr18_68189846_68190022 | 0.13 |
Gm18149 |
predicted gene, 18149 |
20967 |
0.17 |
| chr8_45630758_45630927 | 0.13 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
2641 |
0.27 |
| chr16_20722727_20722878 | 0.13 |
Polr2h |
polymerase (RNA) II (DNA directed) polypeptide H |
4630 |
0.09 |
| chr16_76353569_76353920 | 0.13 |
Nrip1 |
nuclear receptor interacting protein 1 |
19293 |
0.19 |
| chr13_58123386_58123686 | 0.12 |
Hnrnpa0 |
heterogeneous nuclear ribonucleoprotein A0 |
5020 |
0.14 |
| chr6_121225000_121225198 | 0.12 |
Gm15856 |
predicted gene 15856 |
1054 |
0.4 |
| chrX_109505622_109505789 | 0.12 |
Gm4784 |
predicted gene 4784 |
113540 |
0.07 |
| chr3_35885240_35885391 | 0.12 |
Gm7741 |
predicted gene 7741 |
21438 |
0.12 |
| chr14_63181023_63181193 | 0.12 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
1530 |
0.32 |
| chr11_79269272_79269423 | 0.12 |
Gm44787 |
predicted gene 44787 |
4497 |
0.18 |
| chr6_51172952_51173120 | 0.12 |
Mir148a |
microRNA 148a |
96874 |
0.07 |
| chr2_142930084_142930238 | 0.12 |
Kif16bos |
kinesin family member 16B, opposite strand |
27196 |
0.19 |
| chr5_28076725_28077152 | 0.12 |
Insig1 |
insulin induced gene 1 |
2272 |
0.26 |
| chr8_128475036_128475233 | 0.12 |
Nrp1 |
neuropilin 1 |
115737 |
0.06 |
| chr10_81416920_81417099 | 0.12 |
Mir1191b |
microRNA 1191b |
712 |
0.37 |
| chr6_39955337_39955488 | 0.12 |
Gm37995 |
predicted gene, 37995 |
71482 |
0.08 |
| chr10_80758685_80759184 | 0.12 |
Dot1l |
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
3471 |
0.11 |
| chr17_56780034_56780185 | 0.12 |
Rfx2 |
regulatory factor X, 2 (influences HLA class II expression) |
3595 |
0.13 |
| chr9_42261400_42261775 | 0.12 |
Sc5d |
sterol-C5-desaturase |
2613 |
0.25 |
| chr11_60245985_60246136 | 0.12 |
Tom1l2 |
target of myb1-like 2 (chicken) |
391 |
0.78 |
| chr16_31233513_31233746 | 0.11 |
Gm41442 |
predicted gene, 41442 |
39 |
0.96 |
| chr2_92424812_92425012 | 0.11 |
Cry2 |
cryptochrome 2 (photolyase-like) |
7480 |
0.11 |
| chr1_88953578_88953747 | 0.11 |
Gm4753 |
predicted gene 4753 |
21343 |
0.19 |
| chr16_20563601_20563763 | 0.11 |
Gm49744 |
predicted gene, 49744 |
2346 |
0.1 |
| chr2_152361963_152362124 | 0.11 |
Gm14165 |
predicted gene 14165 |
8428 |
0.09 |
| chr10_44447565_44447742 | 0.11 |
Prdm1 |
PR domain containing 1, with ZNF domain |
9557 |
0.19 |
| chr9_53666500_53666950 | 0.11 |
Cul5 |
cullin 5 |
347 |
0.59 |
| chr2_122241045_122241229 | 0.11 |
Sord |
sorbitol dehydrogenase |
6388 |
0.11 |
| chr2_93991476_93991648 | 0.11 |
Gm27027 |
predicted gene, 27027 |
4792 |
0.16 |
| chr18_56974912_56975237 | 0.10 |
C330018D20Rik |
RIKEN cDNA C330018D20 gene |
172 |
0.96 |
| chr10_111170795_111170951 | 0.10 |
Osbpl8 |
oxysterol binding protein-like 8 |
6071 |
0.15 |
| chr18_75599862_75600033 | 0.10 |
Ctif |
CBP80/20-dependent translation initiation factor |
37326 |
0.19 |
| chr10_120015234_120015396 | 0.10 |
Grip1 |
glutamate receptor interacting protein 1 |
22209 |
0.22 |
| chr9_63329385_63329536 | 0.10 |
Map2k5 |
mitogen-activated protein kinase kinase 5 |
26317 |
0.18 |
| chr8_128447394_128447576 | 0.10 |
Nrp1 |
neuropilin 1 |
88088 |
0.08 |
| chr12_83055714_83055884 | 0.10 |
Rgs6 |
regulator of G-protein signaling 6 |
8809 |
0.21 |
| chr7_135655279_135655442 | 0.10 |
5830432E09Rik |
RIKEN cDNA 5830432E09 gene |
3046 |
0.21 |
| chr7_80994654_80994839 | 0.10 |
Gm44967 |
predicted gene 44967 |
63 |
0.92 |
| chrX_74312638_74312789 | 0.10 |
Fam50a |
family with sequence similarity 50, member A |
320 |
0.69 |
| chr9_78139600_78139951 | 0.09 |
Cilk1 |
ciliogenesis associated kinase 1 |
15548 |
0.11 |
| chr15_77125543_77125747 | 0.09 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
7135 |
0.13 |
| chr3_57431501_57431652 | 0.09 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
6262 |
0.24 |
| chr4_35135315_35135501 | 0.09 |
Ifnk |
interferon kappa |
16648 |
0.18 |
| chr9_119159811_119159962 | 0.09 |
Gm47289 |
predicted gene, 47289 |
725 |
0.49 |
| chr16_45836941_45837118 | 0.09 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
7205 |
0.22 |
| chr2_6327142_6327303 | 0.09 |
AL845275.1 |
novel protein |
4142 |
0.21 |
| chr17_28428613_28428954 | 0.09 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr9_122187419_122187656 | 0.09 |
Ano10 |
anoctamin 10 |
82 |
0.96 |
| chr11_51857780_51858123 | 0.09 |
Jade2 |
jade family PHD finger 2 |
298 |
0.9 |
| chr16_43345942_43346096 | 0.09 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
17029 |
0.16 |
| chr10_20233460_20233772 | 0.09 |
Map7 |
microtubule-associated protein 7 |
3821 |
0.18 |
| chr11_22842932_22843203 | 0.08 |
Gm20456 |
predicted gene 20456 |
3768 |
0.14 |
| chr11_98708992_98709189 | 0.08 |
Med24 |
mediator complex subunit 24 |
1087 |
0.28 |
| chr8_104384162_104384337 | 0.08 |
Cmtm4 |
CKLF-like MARVEL transmembrane domain containing 4 |
11558 |
0.09 |
| chr17_30392338_30392489 | 0.08 |
Gm6402 |
predicted pseudogene 6402 |
3620 |
0.22 |
| chr9_86694067_86694240 | 0.08 |
A330041J22Rik |
RIKEN cDNA A330041J22 gene |
1307 |
0.26 |
| chr4_101647476_101647833 | 0.08 |
Leprot |
leptin receptor overlapping transcript |
64 |
0.98 |
| chr4_8696499_8696683 | 0.08 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
5226 |
0.27 |
| chr8_4312939_4313169 | 0.08 |
Elavl1 |
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
435 |
0.72 |
| chr5_146007780_146007932 | 0.08 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
1742 |
0.26 |
| chr11_60878791_60878970 | 0.08 |
Tmem11 |
transmembrane protein 11 |
93 |
0.95 |
| chr4_131918798_131918960 | 0.08 |
Gm12992 |
predicted gene 12992 |
1150 |
0.29 |
| chrX_98205931_98206095 | 0.08 |
Ar |
androgen receptor |
57244 |
0.16 |
| chr19_3891896_3892047 | 0.08 |
Chka |
choline kinase alpha |
240 |
0.83 |
| chr1_189993369_189993532 | 0.08 |
Smyd2 |
SET and MYND domain containing 2 |
71087 |
0.1 |
| chr13_70866767_70867075 | 0.08 |
8030423J24Rik |
RIKEN cDNA 8030423J24 gene |
16027 |
0.18 |
| chr9_57433273_57433508 | 0.07 |
Ppcdc |
phosphopantothenoylcysteine decarboxylase |
1634 |
0.27 |
| chr11_116683735_116683894 | 0.07 |
St6galnac2 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
1002 |
0.33 |
| chr19_40499284_40499488 | 0.07 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
11640 |
0.19 |
| chr3_82022483_82022674 | 0.07 |
Gucy1b1 |
guanylate cyclase 1, soluble, beta 1 |
12744 |
0.17 |
| chr2_118263252_118263413 | 0.07 |
Fsip1 |
fibrous sheath-interacting protein 1 |
6366 |
0.14 |
| chr5_9043907_9044132 | 0.07 |
Gm40264 |
predicted gene, 40264 |
8895 |
0.15 |
| chr10_28649859_28650024 | 0.07 |
Themis |
thymocyte selection associated |
18419 |
0.24 |
| chr1_45869074_45869225 | 0.07 |
Gm5526 |
predicted pseudogene 5526 |
11395 |
0.13 |
| chr4_154161006_154161194 | 0.07 |
Tprgl |
transformation related protein 63 regulated like |
434 |
0.74 |
| chr15_82788072_82788266 | 0.07 |
Gm27618 |
predicted gene, 27618 |
6028 |
0.1 |
| chr12_69766607_69766781 | 0.07 |
Mir681 |
microRNA 681 |
2750 |
0.18 |
| chr4_116026727_116026890 | 0.07 |
Faah |
fatty acid amide hydrolase |
8882 |
0.13 |
| chr15_66297511_66297674 | 0.07 |
Gm27242 |
predicted gene 27242 |
10675 |
0.19 |
| chr1_87472271_87472440 | 0.07 |
Snorc |
secondary ossification center associated regulator of chondrocyte maturation |
184 |
0.92 |
| chr2_160973552_160974077 | 0.07 |
Chd6 |
chromodomain helicase DNA binding protein 6 |
9018 |
0.16 |
| chr17_43644456_43644768 | 0.07 |
Tdrd6 |
tudor domain containing 6 |
14313 |
0.15 |
| chr18_34904259_34904430 | 0.07 |
Etf1 |
eukaryotic translation termination factor 1 |
27663 |
0.1 |
| chr2_18026641_18026818 | 0.07 |
A930004D18Rik |
RIKEN cDNA A930004D18 gene |
2031 |
0.2 |
| chr10_127578533_127578684 | 0.07 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
3277 |
0.14 |
| chr16_97608071_97608276 | 0.07 |
Tmprss2 |
transmembrane protease, serine 2 |
2796 |
0.29 |
| chr3_146564646_146564797 | 0.07 |
Gm36259 |
predicted gene, 36259 |
1452 |
0.31 |
| chr2_168601784_168601950 | 0.07 |
Nfatc2 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
210 |
0.95 |
| chr6_128522304_128522723 | 0.07 |
Pzp |
PZP, alpha-2-macroglobulin like |
4190 |
0.11 |
| chr7_99123439_99123607 | 0.07 |
Gm44975 |
predicted gene 44975 |
64 |
0.96 |
| chr4_53123836_53123987 | 0.06 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
35984 |
0.16 |
| chr16_11457447_11457608 | 0.06 |
Snx29 |
sorting nexin 29 |
10219 |
0.23 |
| chr10_26806204_26806355 | 0.06 |
Arhgap18 |
Rho GTPase activating protein 18 |
16323 |
0.21 |
| chr17_50470292_50470529 | 0.06 |
Plcl2 |
phospholipase C-like 2 |
38993 |
0.2 |
| chr9_64003465_64003638 | 0.06 |
Smad6 |
SMAD family member 6 |
13436 |
0.15 |
| chr16_16213992_16214195 | 0.06 |
Pkp2 |
plakophilin 2 |
775 |
0.67 |
| chr3_145906722_145906888 | 0.06 |
Bcl10 |
B cell leukemia/lymphoma 10 |
15999 |
0.16 |
| chr10_87541894_87542102 | 0.06 |
Pah |
phenylalanine hydroxylase |
4675 |
0.24 |
| chr10_18060923_18061087 | 0.06 |
Reps1 |
RalBP1 associated Eps domain containing protein |
5001 |
0.22 |
| chr13_34232044_34232215 | 0.06 |
Slc22a23 |
solute carrier family 22, member 23 |
38598 |
0.12 |
| chr19_58395752_58395903 | 0.06 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
58639 |
0.13 |
| chr9_43105189_43105381 | 0.06 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
201 |
0.94 |
| chr11_120807985_120808259 | 0.06 |
Fasn |
fatty acid synthase |
483 |
0.62 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |