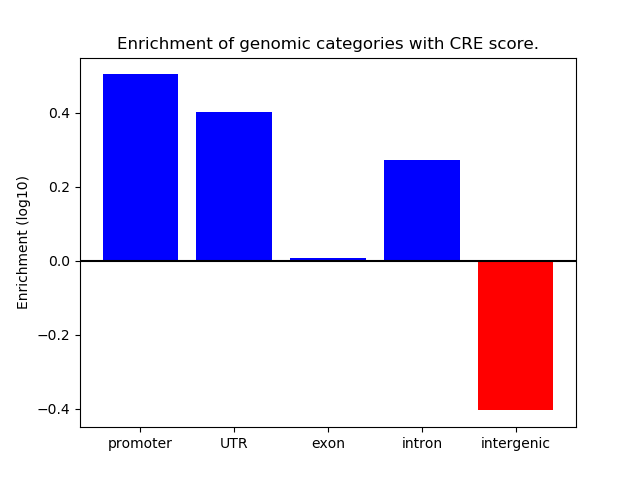
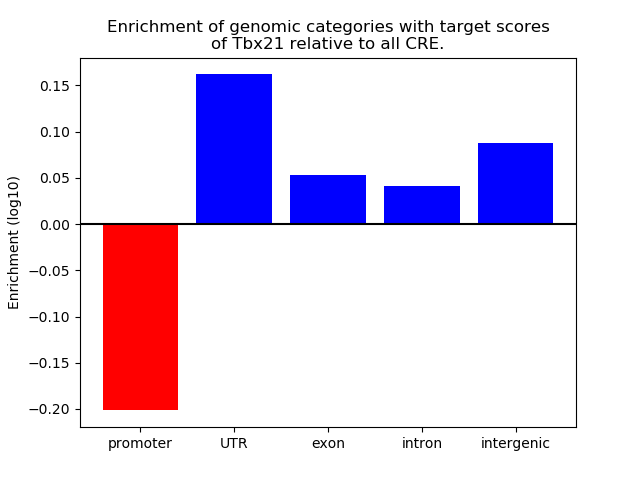
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
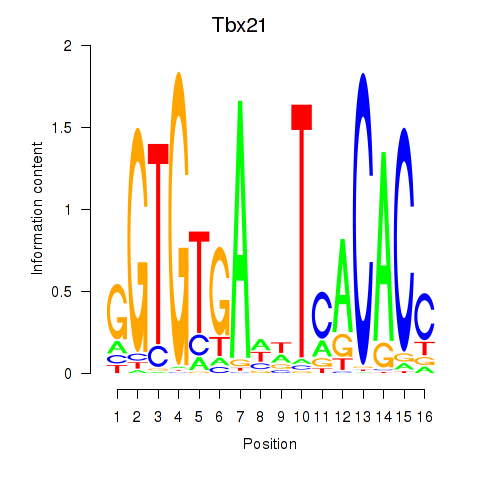
Results for Tbx21
Z-value: 0.66
Transcription factors associated with Tbx21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx21
|
ENSMUSG00000001444.2 | T-box 21 |
Activity of the Tbx21 motif across conditions
Conditions sorted by the z-value of the Tbx21 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
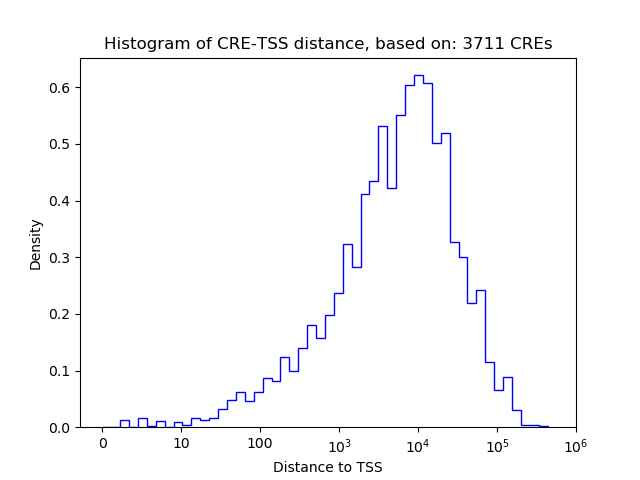
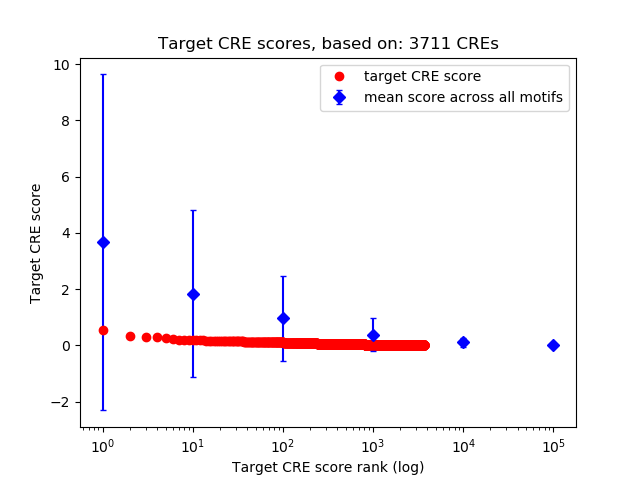
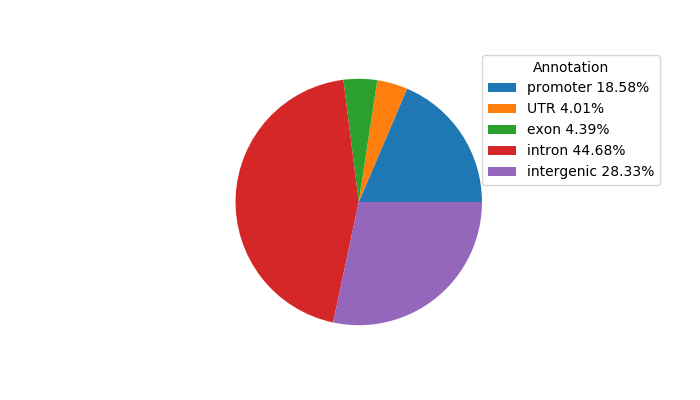
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrY_90740044_90740606 | 0.53 |
Mid1-ps1 |
midline 1, pseudogene 1 |
12732 |
0.18 |
| chr7_24470539_24470701 | 0.34 |
Plaur |
plasminogen activator, urokinase receptor |
8120 |
0.09 |
| chrY_90739770_90739982 | 0.30 |
Mid1-ps1 |
midline 1, pseudogene 1 |
13181 |
0.18 |
| chr9_46116938_46117103 | 0.29 |
Sik3 |
SIK family kinase 3 |
6110 |
0.2 |
| chr11_11848585_11848736 | 0.26 |
Ddc |
dopa decarboxylase |
12380 |
0.17 |
| chr19_47699832_47699983 | 0.23 |
Gm50281 |
predicted gene, 50281 |
3374 |
0.13 |
| chrY_90798307_90798477 | 0.20 |
Gm47283 |
predicted gene, 47283 |
7941 |
0.18 |
| chr5_111659342_111659508 | 0.20 |
Gm42489 |
predicted gene 42489 |
65809 |
0.09 |
| chr7_140851798_140851949 | 0.19 |
Odf3 |
outer dense fiber of sperm tails 3 |
4049 |
0.07 |
| chr12_99371517_99371855 | 0.19 |
Gm47138 |
predicted gene, 47138 |
1773 |
0.26 |
| chr1_55685863_55686014 | 0.19 |
Plcl1 |
phospholipase C-like 1 |
16087 |
0.27 |
| chr11_16832564_16832715 | 0.19 |
Egfros |
epidermal growth factor receptor, opposite strand |
1937 |
0.37 |
| chr6_147838947_147839104 | 0.18 |
Gm15762 |
predicted gene 15762 |
119574 |
0.06 |
| chr1_151215959_151216113 | 0.17 |
Gm47996 |
predicted gene, 47996 |
5905 |
0.13 |
| chr12_10818187_10818338 | 0.17 |
Pgk1-rs7 |
phosphoglycerate kinase-1, related sequence-7 |
81978 |
0.09 |
| chr1_105822945_105823096 | 0.17 |
Tnfrsf11a |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
3150 |
0.26 |
| chr19_44393747_44393961 | 0.16 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12836 |
0.14 |
| chr3_89578871_89579039 | 0.16 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
58791 |
0.09 |
| chr6_97210234_97210388 | 0.16 |
Arl6ip5 |
ADP-ribosylation factor-like 6 interacting protein 5 |
378 |
0.82 |
| chr8_80263585_80263752 | 0.16 |
Gm45430 |
predicted gene 45430 |
68469 |
0.12 |
| chr9_74886235_74886449 | 0.16 |
Onecut1 |
one cut domain, family member 1 |
19858 |
0.14 |
| chr19_28347532_28347702 | 0.16 |
Glis3 |
GLIS family zinc finger 3 |
21611 |
0.26 |
| chr5_125524109_125525122 | 0.15 |
Tmem132b |
transmembrane protein 132B |
7159 |
0.16 |
| chr7_140973288_140973439 | 0.15 |
Ifitm1 |
interferon induced transmembrane protein 1 |
5323 |
0.08 |
| chr8_64812478_64812667 | 0.15 |
Gm10997 |
predicted gene 10997 |
3434 |
0.2 |
| chr5_142438545_142438696 | 0.15 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
25324 |
0.16 |
| chr5_149050829_149051024 | 0.15 |
Hmgb1 |
high mobility group box 1 |
374 |
0.78 |
| chr2_45369277_45369459 | 0.15 |
Gm13479 |
predicted gene 13479 |
12103 |
0.25 |
| chr19_58728798_58728973 | 0.15 |
Pnliprp1 |
pancreatic lipase related protein 1 |
2 |
0.97 |
| chr5_125527806_125528000 | 0.14 |
Tmem132b |
transmembrane protein 132B |
3871 |
0.2 |
| chr1_131741893_131742062 | 0.14 |
Slc26a9 |
solute carrier family 26, member 9 |
2045 |
0.29 |
| chr1_186261473_186261624 | 0.14 |
Gm37272 |
predicted gene, 37272 |
79291 |
0.09 |
| chr7_84596857_84597020 | 0.14 |
Fah |
fumarylacetoacetate hydrolase |
5162 |
0.17 |
| chr9_21634202_21634369 | 0.14 |
Smarca4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
1482 |
0.27 |
| chr3_107608633_107608794 | 0.14 |
Strip1 |
striatin interacting protein 1 |
5368 |
0.15 |
| chr10_119185171_119185322 | 0.14 |
Gm34045 |
predicted gene, 34045 |
18861 |
0.16 |
| chr9_122169762_122169933 | 0.14 |
Snrk |
SNF related kinase |
3103 |
0.18 |
| chr10_95424636_95424797 | 0.14 |
5730420D15Rik |
RIKEN cDNA 5730420D15 gene |
7341 |
0.13 |
| chr15_62131504_62131689 | 0.13 |
Pvt1 |
Pvt1 oncogene |
32567 |
0.22 |
| chr14_73506424_73506580 | 0.13 |
Med4 |
mediator complex subunit 4 |
3523 |
0.2 |
| chr5_32736882_32737075 | 0.13 |
Gm43695 |
predicted gene 43695 |
2356 |
0.17 |
| chr10_99656891_99657084 | 0.13 |
Gm35101 |
predicted gene, 35101 |
12527 |
0.16 |
| chr18_82525967_82526137 | 0.13 |
Rpl21-ps8 |
ribosomal protein L21, pseudogene 8 |
2031 |
0.31 |
| chr1_125968072_125968223 | 0.13 |
Nckap5 |
NCK-associated protein 5 |
2947 |
0.31 |
| chr7_107574446_107574681 | 0.13 |
Olfml1 |
olfactomedin-like 1 |
6797 |
0.16 |
| chr18_76968282_76968441 | 0.13 |
Hdhd2 |
haloacid dehalogenase-like hydrolase domain containing 2 |
23924 |
0.15 |
| chr1_92944221_92944381 | 0.13 |
Capn10 |
calpain 10 |
1597 |
0.22 |
| chr19_3555690_3555841 | 0.13 |
Ppp6r3 |
protein phosphatase 6, regulatory subunit 3 |
19931 |
0.14 |
| chrY_90783982_90784262 | 0.13 |
Gm47283 |
predicted gene, 47283 |
616 |
0.72 |
| chr16_43393519_43393856 | 0.13 |
Gm15713 |
predicted gene 15713 |
26509 |
0.15 |
| chr6_86263585_86263762 | 0.12 |
Gm43917 |
predicted gene, 43917 |
241 |
0.9 |
| chr15_62203372_62203523 | 0.12 |
Pvt1 |
Pvt1 oncogene |
14776 |
0.23 |
| chr11_98772948_98773272 | 0.12 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2223 |
0.16 |
| chr7_126636961_126637113 | 0.12 |
Nupr1 |
nuclear protein transcription regulator 1 |
6176 |
0.08 |
| chr13_103579415_103579566 | 0.12 |
Gm24870 |
predicted gene, 24870 |
62436 |
0.13 |
| chr10_95142612_95142787 | 0.12 |
Gm29684 |
predicted gene, 29684 |
3126 |
0.2 |
| chr6_88003582_88003770 | 0.12 |
4933412L11Rik |
RIKEN cDNA 4933412L11 gene |
9497 |
0.09 |
| chr12_111819925_111820076 | 0.12 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
3744 |
0.14 |
| chr8_27210593_27210748 | 0.12 |
Got1l1 |
glutamic-oxaloacetic transaminase 1-like 1 |
8123 |
0.11 |
| chr12_85299100_85299299 | 0.12 |
Zc2hc1c |
zinc finger, C2HC-type containing 1C |
10608 |
0.1 |
| chr19_16438058_16438255 | 0.12 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
1144 |
0.47 |
| chrX_86300055_86300280 | 0.11 |
Mageb10-ps |
melanoma antigen, family B, 10, pseudogene |
1161 |
0.36 |
| chr1_134071822_134072026 | 0.11 |
Btg2 |
BTG anti-proliferation factor 2 |
7196 |
0.13 |
| chr2_5839169_5839320 | 0.11 |
Cdc123 |
cell division cycle 123 |
5566 |
0.16 |
| chr1_136462619_136462773 | 0.11 |
Kif14 |
kinesin family member 14 |
3647 |
0.18 |
| chr5_138823785_138824155 | 0.11 |
Gm5294 |
predicted gene 5294 |
3890 |
0.23 |
| chr2_36278863_36279014 | 0.11 |
Olfr334-ps1 |
olfactory receptor 334, pseudogene 1 |
6085 |
0.13 |
| chrX_170009083_170009242 | 0.11 |
Erdr1 |
erythroid differentiation regulator 1 |
497 |
0.77 |
| chr13_62870205_62870356 | 0.11 |
Fbp1 |
fructose bisphosphatase 1 |
2478 |
0.2 |
| chr10_71193153_71193343 | 0.11 |
1700113B09Rik |
RIKEN cDNA 1700113B09 gene |
8497 |
0.12 |
| chr6_147638472_147638628 | 0.11 |
1700049E15Rik |
RIKEN cDNA 1700049E15 gene |
51691 |
0.14 |
| chr17_46052426_46052577 | 0.11 |
Vegfa |
vascular endothelial growth factor A |
20132 |
0.13 |
| chr14_65299395_65299546 | 0.11 |
Gm48433 |
predicted gene, 48433 |
26861 |
0.13 |
| chr15_59656049_59656209 | 0.11 |
Trib1 |
tribbles pseudokinase 1 |
7476 |
0.21 |
| chr11_57627460_57627611 | 0.11 |
Galnt10 |
polypeptide N-acetylgalactosaminyltransferase 10 |
17907 |
0.15 |
| chr4_63354324_63354498 | 0.11 |
Orm3 |
orosomucoid 3 |
1751 |
0.21 |
| chr9_60315654_60315805 | 0.11 |
2010001M07Rik |
RIKEN cDNA 2010001M07 gene |
18413 |
0.22 |
| chr14_16479423_16479602 | 0.11 |
Top2b |
topoisomerase (DNA) II beta |
54084 |
0.13 |
| chrX_12189621_12189798 | 0.11 |
Gm14521 |
predicted gene 14521 |
9642 |
0.24 |
| chr18_35345350_35345501 | 0.11 |
Sil1 |
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
2358 |
0.31 |
| chr6_22114158_22114347 | 0.11 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
21294 |
0.25 |
| chr18_9674129_9674299 | 0.11 |
Colec12 |
collectin sub-family member 12 |
33381 |
0.09 |
| chr7_19877096_19877247 | 0.11 |
Gm44659 |
predicted gene 44659 |
5733 |
0.08 |
| chrY_90739212_90739561 | 0.11 |
Mid1-ps1 |
midline 1, pseudogene 1 |
13671 |
0.18 |
| chr7_34212592_34212743 | 0.11 |
Gpi1 |
glucose-6-phosphate isomerase 1 |
769 |
0.44 |
| chr9_74891211_74891440 | 0.10 |
Onecut1 |
one cut domain, family member 1 |
24841 |
0.14 |
| chrX_86250773_86250944 | 0.10 |
Mageb4 |
melanoma antigen, family B, 4 |
5361 |
0.18 |
| chr10_29821551_29821768 | 0.10 |
Gm6390 |
predicted gene 6390 |
40248 |
0.18 |
| chr9_9241537_9241716 | 0.10 |
Arhgap42 |
Rho GTPase activating protein 42 |
2525 |
0.31 |
| chr12_8447841_8448015 | 0.10 |
Gm48076 |
predicted gene, 48076 |
6990 |
0.17 |
| chr7_141373770_141373967 | 0.10 |
B230206H07Rik |
RIKEN cDNA B230206H07 gene |
8758 |
0.07 |
| chr7_80143391_80143542 | 0.10 |
Gm45202 |
predicted gene 45202 |
11703 |
0.11 |
| chr19_44396138_44396537 | 0.10 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
10353 |
0.14 |
| chr1_192078521_192078715 | 0.10 |
Gstp-ps |
glutathione S-transferase, pi, pseudogene |
4168 |
0.15 |
| chr9_21837693_21838006 | 0.10 |
Angptl8 |
angiopoietin-like 8 |
2339 |
0.17 |
| chr4_148427588_148427746 | 0.10 |
Gm23303 |
predicted gene, 23303 |
15818 |
0.13 |
| chr5_102010547_102010959 | 0.10 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
29195 |
0.16 |
| chr5_117104853_117105004 | 0.10 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
527 |
0.72 |
| chr7_122971788_122971939 | 0.10 |
Rbbp6 |
retinoblastoma binding protein 6, ubiquitin ligase |
945 |
0.45 |
| chr9_108847437_108847672 | 0.10 |
Gm23156 |
predicted gene, 23156 |
3076 |
0.12 |
| chr16_58539055_58539227 | 0.10 |
St3gal6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
14898 |
0.16 |
| chr14_64443056_64443273 | 0.10 |
Msra |
methionine sulfoxide reductase A |
2247 |
0.39 |
| chr17_62752618_62752854 | 0.10 |
Efna5 |
ephrin A5 |
128408 |
0.06 |
| chr7_141994235_141994397 | 0.10 |
Brsk2 |
BR serine/threonine kinase 2 |
1106 |
0.35 |
| chr17_28436263_28436419 | 0.10 |
Fkbp5 |
FK506 binding protein 5 |
4669 |
0.12 |
| chr9_61046625_61046784 | 0.10 |
Gm39348 |
predicted gene, 39348 |
8965 |
0.15 |
| chr18_5161126_5161309 | 0.09 |
Gm26682 |
predicted gene, 26682 |
4514 |
0.29 |
| chr9_45395952_45396103 | 0.09 |
Fxyd2 |
FXYD domain-containing ion transport regulator 2 |
3642 |
0.15 |
| chr6_5165732_5165895 | 0.09 |
Pon1 |
paraoxonase 1 |
27950 |
0.16 |
| chr11_16895683_16895855 | 0.09 |
Egfr |
epidermal growth factor receptor |
9416 |
0.2 |
| chr12_36155196_36155406 | 0.09 |
Bzw2 |
basic leucine zipper and W2 domains 2 |
1326 |
0.29 |
| chr13_114809129_114809295 | 0.09 |
Mocs2 |
molybdenum cofactor synthesis 2 |
9024 |
0.27 |
| chr14_7841370_7841604 | 0.09 |
Flnb |
filamin, beta |
23530 |
0.15 |
| chr2_65123593_65123748 | 0.09 |
Cobll1 |
Cobl-like 1 |
17811 |
0.24 |
| chr13_54113905_54114067 | 0.09 |
Sfxn1 |
sideroflexin 1 |
21275 |
0.16 |
| chr11_43960310_43960461 | 0.09 |
Gm12153 |
predicted gene 12153 |
1296 |
0.56 |
| chr9_65599021_65599172 | 0.09 |
Pif1 |
PIF1 5'-to-3' DNA helicase |
11870 |
0.14 |
| chr9_109432883_109433034 | 0.09 |
Gm42494 |
predicted gene 42494 |
7170 |
0.12 |
| chr2_72206740_72207276 | 0.09 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
8209 |
0.18 |
| chr17_15497016_15497194 | 0.09 |
Psmb1 |
proteasome (prosome, macropain) subunit, beta type 1 |
1193 |
0.32 |
| chrX_59751807_59751986 | 0.09 |
Gm44500 |
predicted gene, 44500 |
58347 |
0.14 |
| chr14_99771220_99771379 | 0.09 |
Gm22970 |
predicted gene, 22970 |
2509 |
0.31 |
| chr8_85498842_85499034 | 0.09 |
Gpt2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
6329 |
0.18 |
| chr12_8511284_8511522 | 0.09 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
202 |
0.93 |
| chr6_82362227_82362387 | 0.09 |
Gm44209 |
predicted gene, 44209 |
39324 |
0.16 |
| chr14_78937878_78938029 | 0.09 |
Gm49016 |
predicted gene, 49016 |
9274 |
0.2 |
| chr5_107949724_107949919 | 0.09 |
Dipk1a |
divergent protein kinase domain 1A |
23177 |
0.11 |
| chr6_83107914_83108197 | 0.09 |
Ccdc142os |
coiled-coil domain containing 142, opposite strand |
932 |
0.17 |
| chr8_36269996_36270164 | 0.09 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
20564 |
0.19 |
| chr6_29523942_29524117 | 0.09 |
Irf5 |
interferon regulatory factor 5 |
2596 |
0.16 |
| chr6_83631962_83632132 | 0.09 |
Clec4f |
C-type lectin domain family 4, member f |
24074 |
0.1 |
| chr5_43306323_43306495 | 0.09 |
6030400A10Rik |
RIKEN cDNA 6030400A10 gene |
41252 |
0.11 |
| chr13_54112633_54112796 | 0.09 |
Sfxn1 |
sideroflexin 1 |
20003 |
0.16 |
| chr15_73148841_73149019 | 0.09 |
Ago2 |
argonaute RISC catalytic subunit 2 |
28917 |
0.16 |
| chr5_140369704_140369867 | 0.09 |
Snx8 |
sorting nexin 8 |
2675 |
0.19 |
| chr19_26640809_26640981 | 0.09 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
17440 |
0.19 |
| chr4_35084216_35084391 | 0.09 |
Ifnk |
interferon kappa |
67753 |
0.09 |
| chr12_116739718_116739887 | 0.09 |
Gm48119 |
predicted gene, 48119 |
113300 |
0.06 |
| chr14_63145558_63145861 | 0.09 |
Ctsb |
cathepsin B |
10191 |
0.13 |
| chr1_88654880_88655031 | 0.08 |
Gm29336 |
predicted gene 29336 |
1353 |
0.35 |
| chr7_99153594_99153745 | 0.08 |
Gm45185 |
predicted gene 45185 |
12076 |
0.12 |
| chr7_16656304_16656459 | 0.08 |
Ceacam-ps1 |
carcinoembryonic antigen-related cell adhesion molecule pseudogene 1 |
1095 |
0.31 |
| chr16_38618107_38618290 | 0.08 |
Tmem39a |
transmembrane protein 39a |
30159 |
0.13 |
| chr1_74331641_74331792 | 0.08 |
Pnkd |
paroxysmal nonkinesiogenic dyskinesia |
894 |
0.38 |
| chr10_117884880_117885031 | 0.08 |
Gm32552 |
predicted gene, 32552 |
18240 |
0.14 |
| chr19_16447855_16448017 | 0.08 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
8636 |
0.18 |
| chrX_86251182_86251377 | 0.08 |
Mageb4 |
melanoma antigen, family B, 4 |
4940 |
0.18 |
| chr9_83202676_83202877 | 0.08 |
Gm38279 |
predicted gene, 38279 |
26999 |
0.15 |
| chr9_45299334_45299510 | 0.08 |
Gm47964 |
predicted gene, 47964 |
2540 |
0.16 |
| chr13_9032093_9032260 | 0.08 |
Gtpbp4 |
GTP binding protein 4 |
36093 |
0.09 |
| chr9_69682049_69682208 | 0.08 |
B230323A14Rik |
RIKEN cDNA B230323A14 gene |
76612 |
0.08 |
| chr13_47179143_47179300 | 0.08 |
Rnf144b |
ring finger protein 144B |
14604 |
0.15 |
| chr10_5264488_5264639 | 0.08 |
Syne1 |
spectrin repeat containing, nuclear envelope 1 |
69856 |
0.12 |
| chr3_131299141_131299297 | 0.08 |
4930534D22Rik |
RIKEN cDNA 4930534D22 gene |
3424 |
0.17 |
| chr7_48892680_48892953 | 0.08 |
Gm2788 |
predicted gene 2788 |
5586 |
0.14 |
| chr13_41607994_41608177 | 0.08 |
Tmem170b |
transmembrane protein 170B |
1869 |
0.31 |
| chr1_138322744_138322953 | 0.08 |
Atp6v1g3 |
ATPase, H+ transporting, lysosomal V1 subunit G3 |
49110 |
0.12 |
| chr17_71251888_71252113 | 0.08 |
Emilin2 |
elastin microfibril interfacer 2 |
3406 |
0.2 |
| chr13_41386761_41386912 | 0.08 |
Gm48571 |
predicted gene, 48571 |
6880 |
0.15 |
| chr15_74872740_74872891 | 0.08 |
Ly6m |
lymphocyte antigen 6 complex, locus M |
8889 |
0.1 |
| chr3_146876008_146876164 | 0.08 |
Ttll7 |
tubulin tyrosine ligase-like family, member 7 |
18090 |
0.19 |
| chr11_119007538_119007733 | 0.08 |
Cbx2 |
chromobox 2 |
15327 |
0.15 |
| chr2_73983560_73983744 | 0.08 |
Gm13668 |
predicted gene 13668 |
22983 |
0.2 |
| chr13_104616882_104617070 | 0.08 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
67955 |
0.13 |
| chr12_12904564_12904746 | 0.08 |
4930519A11Rik |
RIKEN cDNA 4930519A11 gene |
105 |
0.95 |
| chr2_18091943_18092094 | 0.08 |
Mllt10 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
27177 |
0.1 |
| chr15_81801453_81801720 | 0.08 |
Tef |
thyrotroph embryonic factor |
835 |
0.36 |
| chr13_46165206_46165365 | 0.08 |
Gm10113 |
predicted gene 10113 |
25761 |
0.22 |
| chr18_81321681_81321954 | 0.08 |
Gm30192 |
predicted gene, 30192 |
57109 |
0.13 |
| chr4_135515022_135515191 | 0.08 |
Gm25317 |
predicted gene, 25317 |
2627 |
0.19 |
| chr14_70381695_70381872 | 0.08 |
Gm9174 |
predicted pseudogene 9174 |
21596 |
0.1 |
| chr1_136141628_136141817 | 0.08 |
Kif21b |
kinesin family member 21B |
10268 |
0.11 |
| chr19_47449301_47449452 | 0.08 |
Sh3pxd2a |
SH3 and PX domains 2A |
14871 |
0.19 |
| chr15_56244131_56244282 | 0.08 |
Hba-ps3 |
hemoglobin alpha, pseudogene 3 |
71817 |
0.12 |
| chr15_57741253_57741423 | 0.07 |
9330154K18Rik |
RIKEN cDNA 9330154K18 gene |
2672 |
0.3 |
| chr11_111444682_111444833 | 0.07 |
Gm11676 |
predicted gene 11676 |
168549 |
0.04 |
| chr9_79876120_79876328 | 0.07 |
Gm3211 |
predicted gene 3211 |
36976 |
0.12 |
| chr8_122517598_122517757 | 0.07 |
Gm26497 |
predicted gene, 26497 |
2997 |
0.13 |
| chr15_28839041_28839211 | 0.07 |
Gm49280 |
predicted gene, 49280 |
15878 |
0.29 |
| chr6_147950991_147951177 | 0.07 |
Far2 |
fatty acyl CoA reductase 2 |
96175 |
0.07 |
| chr8_48729447_48729608 | 0.07 |
Tenm3 |
teneurin transmembrane protein 3 |
54837 |
0.15 |
| chr12_25265696_25265865 | 0.07 |
Gm19340 |
predicted gene, 19340 |
16815 |
0.18 |
| chr11_95738579_95738882 | 0.07 |
Zfp652 |
zinc finger protein 652 |
10270 |
0.13 |
| chr7_35448631_35448822 | 0.07 |
Slc7a9 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
70 |
0.96 |
| chr2_173682841_173683018 | 0.07 |
Mir6340 |
microRNA 6340 |
18219 |
0.13 |
| chr18_69641471_69641660 | 0.07 |
Tcf4 |
transcription factor 4 |
15102 |
0.27 |
| chr5_114152481_114152649 | 0.07 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
6030 |
0.13 |
| chr8_122898814_122898990 | 0.07 |
Ankrd11 |
ankyrin repeat domain 11 |
878 |
0.49 |
| chr3_79528511_79528691 | 0.07 |
Fnip2 |
folliculin interacting protein 2 |
3275 |
0.2 |
| chr11_3417807_3418012 | 0.07 |
Limk2 |
LIM motif-containing protein kinase 2 |
8720 |
0.12 |
| chr1_167305605_167305772 | 0.07 |
Tmco1 |
transmembrane and coiled-coil domains 1 |
2690 |
0.16 |
| chr13_93625401_93625555 | 0.07 |
Gm15622 |
predicted gene 15622 |
96 |
0.96 |
| chr2_144198372_144198743 | 0.07 |
Gm5535 |
predicted gene 5535 |
7311 |
0.15 |
| chr9_57680637_57680986 | 0.07 |
Cyp1a2 |
cytochrome P450, family 1, subfamily a, polypeptide 2 |
1225 |
0.34 |
| chr11_115421469_115421637 | 0.07 |
Kctd2 |
potassium channel tetramerisation domain containing 2 |
1241 |
0.21 |
| chr15_77339159_77339335 | 0.07 |
Gm49436 |
predicted gene, 49436 |
27452 |
0.1 |
| chr10_8518750_8519222 | 0.07 |
Ust |
uronyl-2-sulfotransferase |
161 |
0.97 |
| chr9_37614348_37614562 | 0.07 |
Siae |
sialic acid acetylesterase |
628 |
0.43 |
| chr15_63616907_63617090 | 0.07 |
Gm5473 |
predicted pseudogene 5473 |
14699 |
0.21 |
| chr10_93431595_93431770 | 0.07 |
Mir7688 |
microRNA 7688 |
1425 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |