Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
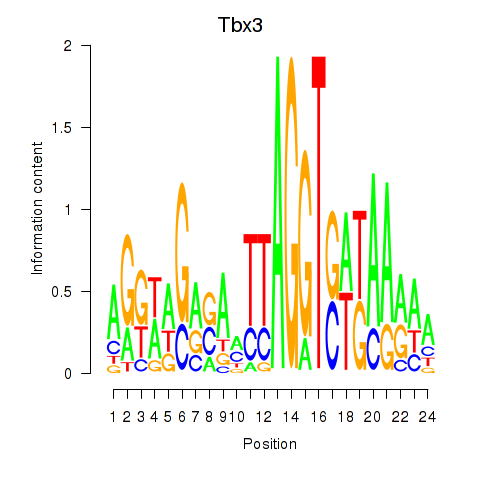
Results for Tbx3
Z-value: 2.60
Transcription factors associated with Tbx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx3
|
ENSMUSG00000018604.12 | T-box 3 |
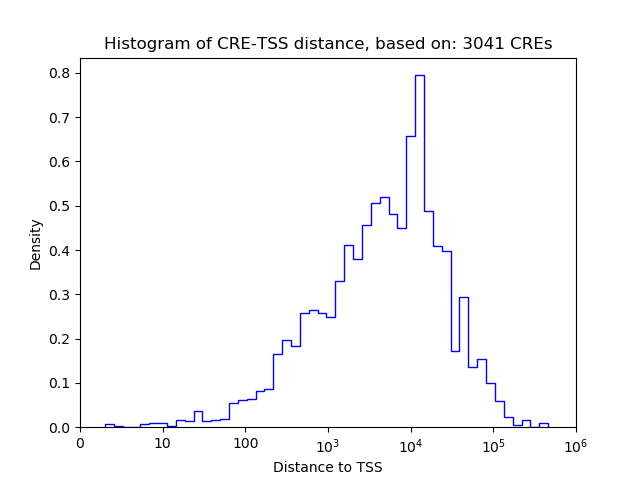
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_119673395_119673565 | Tbx3 | 1965 | 0.230929 | 0.95 | 3.5e-03 | Click! |
| chr5_119670635_119671101 | Tbx3 | 6 | 0.897434 | -0.82 | 4.5e-02 | Click! |
| chr5_119674570_119674817 | Tbx3 | 752 | 0.567953 | 0.79 | 6.1e-02 | Click! |
| chr5_119669855_119670035 | Tbx3 | 724 | 0.521969 | -0.77 | 7.4e-02 | Click! |
| chr5_119674194_119674345 | Tbx3 | 1176 | 0.384983 | 0.67 | 1.5e-01 | Click! |
Activity of the Tbx3 motif across conditions
Conditions sorted by the z-value of the Tbx3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
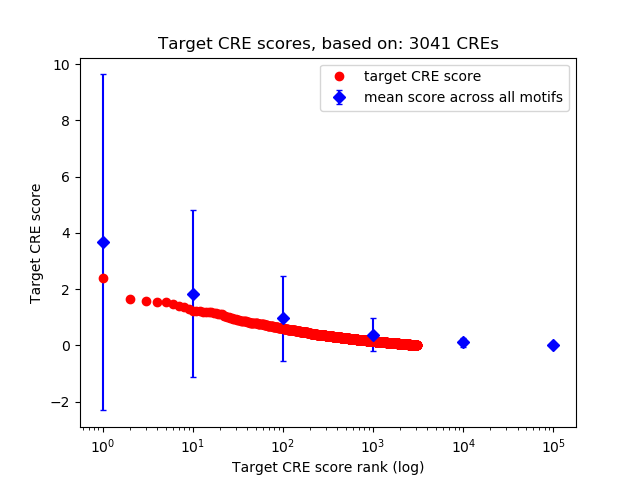
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_10040748_10040899 | 2.39 |
Tnfsf13b |
tumor necrosis factor (ligand) superfamily, member 13b |
9505 |
0.16 |
| chr6_24610042_24610446 | 1.66 |
Lmod2 |
leiomodin 2 (cardiac) |
12482 |
0.14 |
| chr7_113961588_113961904 | 1.57 |
Gm45615 |
predicted gene 45615 |
125152 |
0.05 |
| chrX_97748997_97749155 | 1.55 |
Gm9062 |
predicted gene 9062 |
77303 |
0.12 |
| chr9_122056238_122056617 | 1.53 |
Gm39465 |
predicted gene, 39465 |
4964 |
0.13 |
| chr13_31477486_31477662 | 1.46 |
Gm23351 |
predicted gene, 23351 |
50319 |
0.1 |
| chr2_70813245_70813396 | 1.41 |
Tlk1 |
tousled-like kinase 1 |
11908 |
0.21 |
| chr8_93180612_93180763 | 1.35 |
Ces1d |
carboxylesterase 1D |
5398 |
0.15 |
| chr8_26982042_26982193 | 1.28 |
Gm45371 |
predicted gene 45371 |
2116 |
0.17 |
| chr15_3459919_3460070 | 1.23 |
Ghr |
growth hormone receptor |
11650 |
0.28 |
| chr1_182500136_182500458 | 1.22 |
Gm37069 |
predicted gene, 37069 |
6141 |
0.15 |
| chr16_23112995_23113176 | 1.20 |
Gm24616 |
predicted gene, 24616 |
1468 |
0.12 |
| chr16_25086305_25086484 | 1.20 |
A230028O05Rik |
RIKEN cDNA A230028O05 gene |
26755 |
0.26 |
| chr13_41202938_41203089 | 1.20 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
17149 |
0.11 |
| chr18_33780011_33780201 | 1.18 |
Epb41l4aos |
erythrocyte membrane protein band 4.1 like 4a, opposite strand |
14786 |
0.18 |
| chr3_97639903_97640190 | 1.17 |
Fmo5 |
flavin containing monooxygenase 5 |
11163 |
0.13 |
| chr4_109800666_109800954 | 1.16 |
Faf1 |
Fas-associated factor 1 |
39975 |
0.16 |
| chr11_106526874_106527175 | 1.15 |
Gm22711 |
predicted gene, 22711 |
25779 |
0.13 |
| chr1_102517902_102518283 | 1.13 |
Gm20281 |
predicted gene, 20281 |
59170 |
0.14 |
| chr6_134461351_134461502 | 1.11 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
3582 |
0.23 |
| chr11_43927784_43928040 | 1.11 |
Adra1b |
adrenergic receptor, alpha 1b |
26702 |
0.2 |
| chr2_85071327_85071510 | 1.04 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
10454 |
0.12 |
| chr9_27802459_27802634 | 1.03 |
Opcml |
opioid binding protein/cell adhesion molecule-like |
11244 |
0.3 |
| chr1_13287819_13287974 | 1.00 |
Gm38223 |
predicted gene, 38223 |
2610 |
0.18 |
| chr18_11144853_11145048 | 0.99 |
Gata6 |
GATA binding protein 6 |
85903 |
0.09 |
| chr10_8951017_8951174 | 0.99 |
Gm48728 |
predicted gene, 48728 |
854 |
0.63 |
| chr7_29459356_29459516 | 0.98 |
Sipa1l3 |
signal-induced proliferation-associated 1 like 3 |
46016 |
0.11 |
| chr19_59344610_59345004 | 0.94 |
Pdzd8 |
PDZ domain containing 8 |
973 |
0.48 |
| chr5_77312121_77312319 | 0.92 |
Polr2b |
polymerase (RNA) II (DNA directed) polypeptide B |
1729 |
0.23 |
| chr9_111261486_111261656 | 0.92 |
Mlh1 |
mutL homolog 1 |
28 |
0.97 |
| chr8_41194427_41194625 | 0.92 |
Fgl1 |
fibrinogen-like protein 1 |
20627 |
0.16 |
| chr4_155137056_155137330 | 0.91 |
Morn1 |
MORN repeat containing 1 |
26503 |
0.13 |
| chr3_52296364_52296605 | 0.89 |
Gm38034 |
predicted gene, 38034 |
14243 |
0.14 |
| chr17_49278885_49279043 | 0.88 |
Gm22978 |
predicted gene, 22978 |
13778 |
0.2 |
| chr8_22877403_22877583 | 0.88 |
Gm45555 |
predicted gene 45555 |
3904 |
0.19 |
| chr14_12084762_12084930 | 0.87 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
6314 |
0.26 |
| chr3_94702618_94702790 | 0.86 |
Selenbp2 |
selenium binding protein 2 |
9045 |
0.11 |
| chr7_142720846_142721002 | 0.86 |
Ins2 |
insulin II |
21414 |
0.12 |
| chr13_73604421_73604849 | 0.85 |
Clptm1l |
CLPTM1-like |
629 |
0.7 |
| chr14_63236075_63236286 | 0.85 |
Gata4 |
GATA binding protein 4 |
9068 |
0.17 |
| chr1_159269140_159269294 | 0.83 |
Cop1 |
COP1, E3 ubiquitin ligase |
2483 |
0.26 |
| chr6_144781221_144781581 | 0.82 |
Sox5 |
SRY (sex determining region Y)-box 5 |
518 |
0.79 |
| chr7_55910853_55911161 | 0.81 |
Cyfip1 |
cytoplasmic FMR1 interacting protein 1 |
2635 |
0.18 |
| chr2_25628694_25629041 | 0.81 |
Fcna |
ficolin A |
837 |
0.28 |
| chr6_31261904_31262196 | 0.80 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
15798 |
0.16 |
| chr12_51817932_51818125 | 0.80 |
Hectd1 |
HECT domain E3 ubiquitin protein ligase 1 |
11290 |
0.19 |
| chr9_44946661_44946812 | 0.80 |
Gm19121 |
predicted gene, 19121 |
8979 |
0.08 |
| chr16_93875336_93875598 | 0.79 |
Chaf1b |
chromatin assembly factor 1, subunit B (p60) |
8434 |
0.13 |
| chr13_59825892_59826043 | 0.79 |
Gm34961 |
predicted gene, 34961 |
2701 |
0.15 |
| chr9_55240591_55240817 | 0.79 |
Nrg4 |
neuregulin 4 |
1757 |
0.34 |
| chr10_75860456_75860626 | 0.79 |
Mif |
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
301 |
0.8 |
| chr5_3599098_3599294 | 0.78 |
Rbm48 |
RNA binding motif protein 48 |
2611 |
0.12 |
| chr18_64626918_64627221 | 0.77 |
Gm6978 |
predicted gene 6978 |
13970 |
0.14 |
| chr2_118263441_118263625 | 0.77 |
Fsip1 |
fibrous sheath-interacting protein 1 |
6567 |
0.14 |
| chr5_122312662_122313034 | 0.77 |
Gm15842 |
predicted gene 15842 |
10426 |
0.1 |
| chrX_13047364_13047515 | 0.77 |
5730405O15Rik |
RIKEN cDNA 5730405O15 gene |
5388 |
0.21 |
| chr13_93698699_93698858 | 0.76 |
Dmgdh |
dimethylglycine dehydrogenase precursor |
12761 |
0.13 |
| chr9_106264996_106265377 | 0.76 |
Poc1a |
POC1 centriolar protein A |
15875 |
0.1 |
| chr3_95214280_95214549 | 0.75 |
Gm16740 |
predicted gene, 16740 |
3043 |
0.1 |
| chr10_89471569_89471720 | 0.75 |
Gas2l3 |
growth arrest-specific 2 like 3 |
27677 |
0.18 |
| chr17_47976252_47976712 | 0.74 |
Gm14871 |
predicted gene 14871 |
27090 |
0.11 |
| chr6_51635883_51636035 | 0.74 |
Gm22914 |
predicted gene, 22914 |
14327 |
0.23 |
| chr3_79178795_79179141 | 0.73 |
Rapgef2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
2372 |
0.3 |
| chr6_50037983_50038427 | 0.72 |
Gm3455 |
predicted gene 3455 |
12981 |
0.26 |
| chr1_9743510_9743661 | 0.71 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
4063 |
0.17 |
| chr16_18074643_18074794 | 0.71 |
Prodh |
proline dehydrogenase |
4504 |
0.15 |
| chr4_148903259_148903418 | 0.71 |
Casz1 |
castor zinc finger 1 |
2189 |
0.28 |
| chr6_122636931_122637082 | 0.70 |
Dppa3 |
developmental pluripotency-associated 3 |
10253 |
0.12 |
| chr10_89627085_89627236 | 0.70 |
Slc17a8 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
5907 |
0.2 |
| chr11_80974682_80974867 | 0.70 |
Gm11416 |
predicted gene 11416 |
72020 |
0.1 |
| chr6_85835048_85835221 | 0.69 |
Nat8 |
N-acetyltransferase 8 (GCN5-related) |
3052 |
0.12 |
| chr3_145823776_145824250 | 0.69 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
13534 |
0.2 |
| chr9_19691303_19691463 | 0.68 |
Gm7769 |
predicted gene 7769 |
764 |
0.54 |
| chr17_64763760_64763987 | 0.68 |
Dreh |
down-regulated in hepatocellular carcinoma |
3238 |
0.25 |
| chr6_90508904_90509121 | 0.68 |
Gm44236 |
predicted gene, 44236 |
4290 |
0.14 |
| chr2_147961290_147961525 | 0.67 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
14778 |
0.22 |
| chr2_117250087_117250471 | 0.67 |
Fam98b |
family with sequence similarity 98, member B |
540 |
0.71 |
| chr7_67662739_67663027 | 0.66 |
Ttc23 |
tetratricopeptide repeat domain 23 |
3905 |
0.16 |
| chr12_87895766_87895919 | 0.66 |
Gm2022 |
predicted pseudogene 2022 |
472 |
0.56 |
| chr1_107939897_107940125 | 0.66 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
3302 |
0.23 |
| chr16_38246776_38247000 | 0.65 |
Gsk3b |
glycogen synthase kinase 3 beta |
38850 |
0.11 |
| chr13_101350915_101351127 | 0.65 |
Gm2534 |
predicted gene 2534 |
2269 |
0.35 |
| chr10_59476612_59476997 | 0.65 |
Mcu |
mitochondrial calcium uniporter |
18644 |
0.18 |
| chr17_26588858_26589009 | 0.64 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
5943 |
0.12 |
| chr17_12941646_12941826 | 0.64 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
1136 |
0.27 |
| chr2_91241235_91241386 | 0.64 |
Ddb2 |
damage specific DNA binding protein 2 |
4328 |
0.12 |
| chr9_70654222_70654379 | 0.64 |
Gm10642 |
predicted gene 10642 |
1951 |
0.28 |
| chr4_82465338_82465490 | 0.64 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
26004 |
0.18 |
| chr2_71633528_71633687 | 0.63 |
Platr26 |
pluripotency associated transcript 26 |
85810 |
0.06 |
| chr2_177470070_177470297 | 0.62 |
Zfp970 |
zinc finger protein 970 |
5337 |
0.17 |
| chr8_109995594_109996260 | 0.62 |
Tat |
tyrosine aminotransferase |
5421 |
0.13 |
| chr3_97616813_97616964 | 0.62 |
Chd1l |
chromodomain helicase DNA binding protein 1-like |
6685 |
0.15 |
| chr6_31260950_31261295 | 0.62 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
16726 |
0.15 |
| chr13_93627456_93627944 | 0.61 |
Gm15622 |
predicted gene 15622 |
2318 |
0.25 |
| chr9_74530860_74531022 | 0.61 |
Gm28622 |
predicted gene 28622 |
30437 |
0.19 |
| chr13_63291371_63291534 | 0.61 |
Aopep |
aminopeptidase O |
7341 |
0.08 |
| chr6_146926763_146926939 | 0.61 |
1700034J05Rik |
RIKEN cDNA 1700034J05 gene |
27563 |
0.13 |
| chr14_61687453_61687640 | 0.60 |
Gm37820 |
predicted gene, 37820 |
4036 |
0.12 |
| chr2_160805916_160806220 | 0.60 |
Gm11447 |
predicted gene 11447 |
39545 |
0.11 |
| chr16_52336165_52336365 | 0.60 |
Alcam |
activated leukocyte cell adhesion molecule |
39341 |
0.22 |
| chr3_101572198_101572792 | 0.60 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
5065 |
0.19 |
| chr15_10916982_10917286 | 0.60 |
Gm29742 |
predicted gene, 29742 |
303 |
0.89 |
| chr2_91636079_91636258 | 0.59 |
F2 |
coagulation factor II |
227 |
0.88 |
| chr2_27470919_27471184 | 0.59 |
Brd3 |
bromodomain containing 3 |
4130 |
0.16 |
| chr11_54437742_54438063 | 0.58 |
Fnip1 |
folliculin interacting protein 1 |
297 |
0.87 |
| chr3_95323328_95323494 | 0.58 |
Cers2 |
ceramide synthase 2 |
2471 |
0.13 |
| chr4_63217231_63217382 | 0.57 |
Col27a1 |
collagen, type XXVII, alpha 1 |
1871 |
0.32 |
| chr19_44399803_44399962 | 0.57 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
6808 |
0.15 |
| chr9_116174486_116174663 | 0.57 |
Tgfbr2 |
transforming growth factor, beta receptor II |
691 |
0.68 |
| chr2_122248835_122249017 | 0.57 |
Sord |
sorbitol dehydrogenase |
14177 |
0.1 |
| chr1_179805658_179805822 | 0.57 |
Ahctf1 |
AT hook containing transcription factor 1 |
2060 |
0.29 |
| chr6_50132856_50133185 | 0.57 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
22160 |
0.22 |
| chr1_160786063_160786275 | 0.56 |
Rabgap1l |
RAB GTPase activating protein 1-like |
6769 |
0.13 |
| chr3_58685911_58686282 | 0.56 |
Siah2 |
siah E3 ubiquitin protein ligase 2 |
59 |
0.96 |
| chr6_36174952_36175118 | 0.56 |
Gm43443 |
predicted gene 43443 |
14064 |
0.28 |
| chr10_61666021_61666218 | 0.56 |
Ppa1 |
pyrophosphatase (inorganic) 1 |
2430 |
0.19 |
| chr18_66004496_66004733 | 0.55 |
Lman1 |
lectin, mannose-binding, 1 |
1975 |
0.25 |
| chr13_50595486_50595664 | 0.55 |
Mir683-2 |
microRNA 683-2 |
5505 |
0.18 |
| chr5_30248189_30248430 | 0.55 |
Gm16288 |
predicted gene 16288 |
7861 |
0.14 |
| chr8_18711435_18711873 | 0.55 |
Angpt2 |
angiopoietin 2 |
12107 |
0.18 |
| chr1_133374276_133374730 | 0.55 |
Etnk2 |
ethanolamine kinase 2 |
7216 |
0.14 |
| chr13_17838169_17838320 | 0.55 |
Gm18859 |
predicted gene, 18859 |
17298 |
0.12 |
| chr19_25090050_25090416 | 0.54 |
Dock8 |
dedicator of cytokinesis 8 |
12319 |
0.24 |
| chr8_3180016_3180192 | 0.54 |
Insr |
insulin receptor |
1747 |
0.36 |
| chr6_136554845_136555019 | 0.54 |
Atf7ip |
activating transcription factor 7 interacting protein |
910 |
0.54 |
| chr15_58941890_58942087 | 0.54 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
8180 |
0.13 |
| chr6_149155690_149155853 | 0.54 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
14125 |
0.12 |
| chr13_119418806_119418992 | 0.54 |
Paip1 |
polyadenylate binding protein-interacting protein 1 |
9702 |
0.16 |
| chr13_9014068_9014362 | 0.54 |
Gtpbp4 |
GTP binding protein 4 |
18132 |
0.1 |
| chr15_66611144_66611320 | 0.54 |
Phf20l1 |
PHD finger protein 20-like 1 |
2862 |
0.3 |
| chr11_60017161_60017312 | 0.54 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
3990 |
0.19 |
| chr17_31390793_31390965 | 0.54 |
Pde9a |
phosphodiesterase 9A |
4585 |
0.17 |
| chr15_39150023_39150210 | 0.54 |
n-R5s39 |
nuclear encoded rRNA 5S 39 |
3629 |
0.17 |
| chr2_160367492_160367704 | 0.53 |
Mafb |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
533 |
0.84 |
| chr1_92665288_92665439 | 0.53 |
Otos |
otospiralin |
16522 |
0.11 |
| chr14_28996935_28997118 | 0.53 |
Gm23999 |
predicted gene, 23999 |
12773 |
0.19 |
| chr3_107985675_107985968 | 0.53 |
Gstm2 |
glutathione S-transferase, mu 2 |
539 |
0.52 |
| chr1_155799954_155800105 | 0.52 |
Qsox1 |
quiescin Q6 sulfhydryl oxidase 1 |
4612 |
0.14 |
| chr5_20589642_20589807 | 0.52 |
Gm3544 |
predicted gene 3544 |
1128 |
0.56 |
| chr2_90955947_90956098 | 0.52 |
Celf1 |
CUGBP, Elav-like family member 1 |
9070 |
0.12 |
| chr14_65971926_65972160 | 0.52 |
Clu |
clusterin |
740 |
0.6 |
| chr4_122935428_122935617 | 0.52 |
Gm12891 |
predicted gene 12891 |
9689 |
0.14 |
| chr11_99006922_99007084 | 0.52 |
Top2a |
topoisomerase (DNA) II alpha |
481 |
0.68 |
| chr13_77708354_77708514 | 0.51 |
Fam172a |
family with sequence similarity 172, member A |
256 |
0.93 |
| chr18_61424158_61424549 | 0.51 |
Gm8755 |
predicted gene 8755 |
9848 |
0.12 |
| chr2_61712095_61712270 | 0.50 |
Psmd14 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
391 |
0.85 |
| chr16_45378647_45378817 | 0.50 |
Cd200 |
CD200 antigen |
21580 |
0.16 |
| chr18_12638986_12639155 | 0.50 |
Ttc39c |
tetratricopeptide repeat domain 39C |
4274 |
0.17 |
| chrY_90839675_90839839 | 0.50 |
Gm21748 |
predicted gene, 21748 |
580 |
0.73 |
| chr10_5409182_5409553 | 0.50 |
Syne1 |
spectrin repeat containing, nuclear envelope 1 |
40881 |
0.18 |
| chr6_6204143_6204472 | 0.50 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
12811 |
0.23 |
| chr1_191072716_191073064 | 0.49 |
Gm38188 |
predicted gene, 38188 |
5790 |
0.11 |
| chr8_105152387_105152546 | 0.49 |
Gm3830 |
predicted gene 3830 |
12121 |
0.09 |
| chr1_68720880_68721031 | 0.49 |
Gm37769 |
predicted gene, 37769 |
7550 |
0.25 |
| chr9_90263339_90263559 | 0.49 |
Tbc1d2b |
TBC1 domain family, member 2B |
7320 |
0.18 |
| chr5_17419608_17419767 | 0.49 |
Gm21999 |
predicted gene 21999 |
39114 |
0.15 |
| chr13_60709000_60709161 | 0.49 |
Dapk1 |
death associated protein kinase 1 |
12667 |
0.17 |
| chr16_78261377_78261547 | 0.49 |
E330011O21Rik |
RIKEN cDNA E330011O21 gene |
4490 |
0.18 |
| chr16_48623561_48623712 | 0.49 |
Morc1 |
microrchidia 1 |
28969 |
0.22 |
| chr1_13639776_13639929 | 0.49 |
Lactb2 |
lactamase, beta 2 |
1441 |
0.37 |
| chr10_95941668_95942041 | 0.49 |
Eea1 |
early endosome antigen 1 |
1097 |
0.49 |
| chr11_99105082_99105257 | 0.49 |
Tns4 |
tensin 4 |
15863 |
0.14 |
| chr5_106467074_106467543 | 0.48 |
Gm26872 |
predicted gene, 26872 |
3699 |
0.21 |
| chrY_90755303_90755481 | 0.48 |
Gm21860 |
predicted gene, 21860 |
75 |
0.97 |
| chr14_22808144_22808302 | 0.48 |
Gm7473 |
predicted gene 7473 |
32979 |
0.23 |
| chr10_86339517_86339668 | 0.48 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
36738 |
0.16 |
| chr7_90138739_90138890 | 0.48 |
Gm45223 |
predicted gene 45223 |
6322 |
0.12 |
| chr8_82255533_82255726 | 0.48 |
Gm38590 |
predicted gene, 38590 |
48094 |
0.15 |
| chr8_23166783_23167138 | 0.47 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
13267 |
0.09 |
| chr10_89624588_89625080 | 0.47 |
Slc17a8 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
3581 |
0.24 |
| chr18_39593242_39593447 | 0.47 |
Gm30961 |
predicted gene, 30961 |
43062 |
0.16 |
| chrX_142477976_142478290 | 0.47 |
Gm25915 |
predicted gene, 25915 |
7314 |
0.19 |
| chr2_157201501_157201807 | 0.47 |
Rbl1 |
RB transcriptional corepressor like 1 |
2858 |
0.22 |
| chr7_63922331_63923024 | 0.47 |
Klf13 |
Kruppel-like factor 13 |
2193 |
0.22 |
| chr1_140017114_140017477 | 0.47 |
Gm26048 |
predicted gene, 26048 |
13470 |
0.15 |
| chr8_35411222_35411415 | 0.47 |
Gm45301 |
predicted gene 45301 |
1812 |
0.31 |
| chr8_84937365_84937533 | 0.47 |
Mast1 |
microtubule associated serine/threonine kinase 1 |
90 |
0.91 |
| chr6_144070883_144071211 | 0.46 |
Sox5os1 |
SRY (sex determining region Y)-box 5, opposite strand 1 |
49139 |
0.17 |
| chr4_108044826_108045169 | 0.46 |
Scp2 |
sterol carrier protein 2, liver |
2882 |
0.17 |
| chr2_69467066_69467239 | 0.46 |
Lrp2 |
low density lipoprotein receptor-related protein 2 |
41993 |
0.15 |
| chr4_118427849_118428470 | 0.46 |
Elovl1 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
15 |
0.95 |
| chr12_33430163_33430314 | 0.46 |
Twistnb |
twist basic helix-loop-helix transcription factor 1 neighbor |
169 |
0.94 |
| chr8_36290613_36290918 | 0.46 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
41249 |
0.14 |
| chr13_98896782_98896967 | 0.46 |
Tnpo1 |
transportin 1 |
5834 |
0.15 |
| chr8_95358911_95359094 | 0.46 |
Mmp15 |
matrix metallopeptidase 15 |
6734 |
0.13 |
| chr8_36363144_36363313 | 0.45 |
Gm5787 |
predicted gene 5787 |
10405 |
0.21 |
| chr7_67648192_67648343 | 0.45 |
Ttc23 |
tetratricopeptide repeat domain 23 |
807 |
0.52 |
| chr1_195230186_195230410 | 0.45 |
Gm24802 |
predicted gene, 24802 |
2020 |
0.28 |
| chr11_69094794_69095306 | 0.45 |
Per1 |
period circadian clock 1 |
167 |
0.86 |
| chr14_122876950_122877397 | 0.44 |
Pcca |
propionyl-Coenzyme A carboxylase, alpha polypeptide |
1238 |
0.47 |
| chr1_107915528_107915771 | 0.44 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
1610 |
0.39 |
| chr8_122294659_122294822 | 0.44 |
Zfpm1 |
zinc finger protein, multitype 1 |
12580 |
0.14 |
| chr6_59422701_59422852 | 0.44 |
Gprin3 |
GPRIN family member 3 |
3518 |
0.37 |
| chr7_101341850_101342021 | 0.44 |
Stard10 |
START domain containing 10 |
567 |
0.58 |
| chr2_181276195_181276365 | 0.43 |
Gmeb2 |
glucocorticoid modulatory element binding protein 2 |
1802 |
0.2 |
| chr4_105129571_105129783 | 0.43 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
19787 |
0.22 |
| chr9_43222435_43222793 | 0.43 |
Oaf |
out at first homolog |
1360 |
0.36 |
| chr11_76655176_76655342 | 0.43 |
Bhlha9 |
basic helix-loop-helix family, member a9 |
17211 |
0.16 |
| chr15_3420678_3420829 | 0.43 |
Ghr |
growth hormone receptor |
50891 |
0.16 |
| chr2_155587050_155587219 | 0.43 |
Gss |
glutathione synthetase |
442 |
0.63 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.2 | 0.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 0.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.7 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.2 | 0.5 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.2 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 0.5 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.6 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.6 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 0.4 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.4 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.4 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.3 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.1 | 0.5 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 0.3 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.4 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.1 | 0.2 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.1 | 0.2 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.2 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.3 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.4 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.4 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.1 | 0.2 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.3 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.1 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.2 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.3 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.4 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.2 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.2 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.3 | GO:0036336 | dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:1901525 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) establishment of T cell polarity(GO:0001768) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0050932 | regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0070197 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.0 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.8 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.2 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.6 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0034892 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.5 | GO:0016749 | N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0052770 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.6 | GO:0046920 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.0 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |