Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
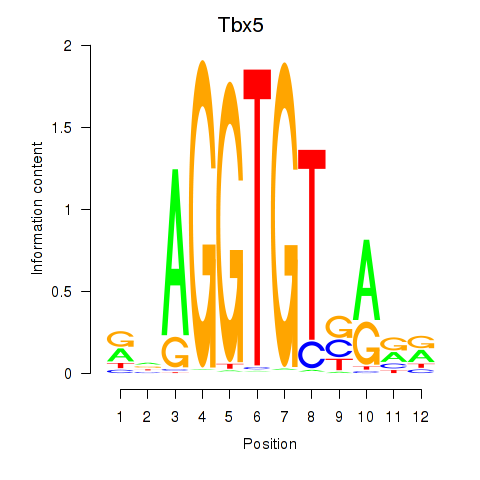
Results for Tbx5
Z-value: 0.79
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSMUSG00000018263.8 | T-box 5 |
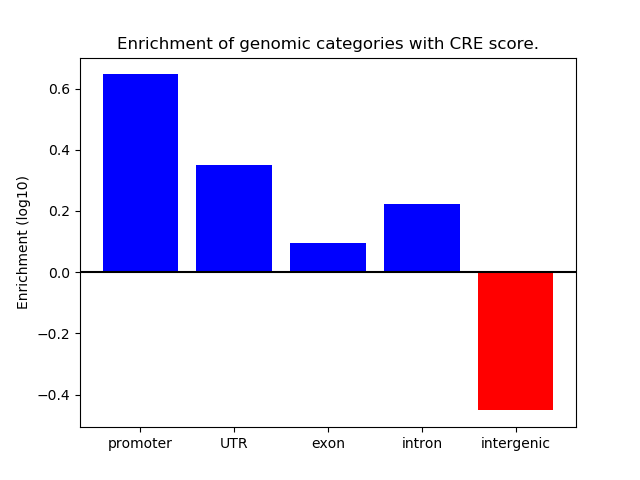
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_119839110_119839360 | Tbx5 | 3080 | 0.227585 | -0.16 | 7.6e-01 | Click! |
Activity of the Tbx5 motif across conditions
Conditions sorted by the z-value of the Tbx5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
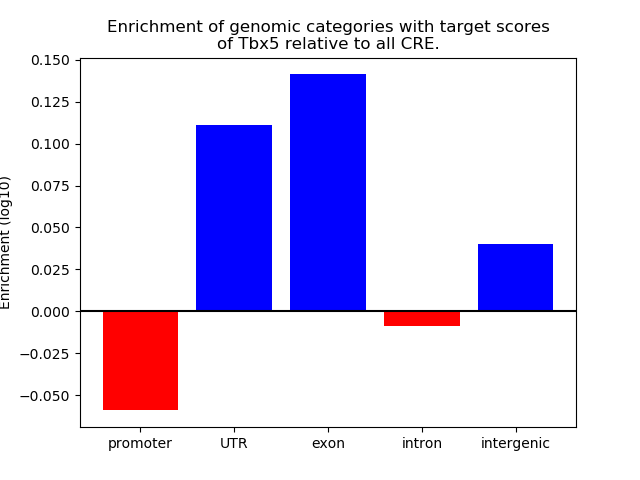
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_31517780_31518546 | 0.34 |
Ass1 |
argininosuccinate synthetase 1 |
327 |
0.88 |
| chr10_117649861_117650065 | 0.30 |
Gm25709 |
predicted gene, 25709 |
12425 |
0.13 |
| chr4_117116805_117116956 | 0.30 |
Ptch2 |
patched 2 |
3456 |
0.09 |
| chrX_169982545_169982696 | 0.29 |
Gm15726 |
predicted gene 15726 |
2612 |
0.19 |
| chr17_45873270_45873443 | 0.28 |
4930542M03Rik |
RIKEN cDNA 4930542M03 gene |
8096 |
0.17 |
| chr3_83043842_83044694 | 0.26 |
Fgb |
fibrinogen beta chain |
5595 |
0.15 |
| chr7_89699637_89700074 | 0.25 |
Me3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
36876 |
0.14 |
| chr3_51254730_51254881 | 0.22 |
Elf2 |
E74-like factor 2 |
5436 |
0.14 |
| chr8_119416779_119416930 | 0.21 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
17270 |
0.13 |
| chr8_119417171_119417322 | 0.21 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
16878 |
0.13 |
| chr3_28025480_28025631 | 0.21 |
Pld1 |
phospholipase D1 |
5707 |
0.3 |
| chr7_13007324_13007475 | 0.21 |
Zbtb45 |
zinc finger and BTB domain containing 45 |
2401 |
0.12 |
| chr12_109798285_109798857 | 0.21 |
Gm9661 |
predicted gene 9661 |
28432 |
0.03 |
| chr2_26314444_26314595 | 0.20 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
996 |
0.38 |
| chr16_25221689_25221840 | 0.20 |
Tprg |
transformation related protein 63 regulated |
65053 |
0.14 |
| chr19_4033739_4033961 | 0.19 |
Gstp1 |
glutathione S-transferase, pi 1 |
2143 |
0.11 |
| chr3_100892764_100892976 | 0.19 |
Trim45 |
tripartite motif-containing 45 |
29332 |
0.16 |
| chr9_98424555_98424727 | 0.19 |
Rbp1 |
retinol binding protein 1, cellular |
1680 |
0.4 |
| chr8_46483334_46483691 | 0.19 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
7454 |
0.16 |
| chr18_63901771_63901922 | 0.18 |
Gm19076 |
predicted gene, 19076 |
2815 |
0.29 |
| chr2_31519156_31519420 | 0.18 |
Ass1 |
argininosuccinate synthetase 1 |
798 |
0.61 |
| chr11_83740503_83740922 | 0.18 |
Wfdc21 |
WAP four-disulfide core domain 21 |
6228 |
0.1 |
| chr8_12798830_12798981 | 0.18 |
Atp11a |
ATPase, class VI, type 11A |
12511 |
0.15 |
| chrX_140539340_140539749 | 0.18 |
Tsc22d3 |
TSC22 domain family, member 3 |
3124 |
0.26 |
| chr11_118454196_118454347 | 0.17 |
Gm11747 |
predicted gene 11747 |
724 |
0.56 |
| chr2_25095172_25095346 | 0.17 |
Noxa1 |
NADPH oxidase activator 1 |
110 |
0.92 |
| chr11_16881299_16881793 | 0.17 |
Egfr |
epidermal growth factor receptor |
3396 |
0.26 |
| chr9_98423199_98423389 | 0.17 |
Rbp1 |
retinol binding protein 1, cellular |
333 |
0.9 |
| chr5_139822534_139822685 | 0.17 |
Tmem184a |
transmembrane protein 184a |
2692 |
0.17 |
| chr9_85551927_85552078 | 0.17 |
Gm48834 |
predicted gene, 48834 |
1763 |
0.42 |
| chr15_88866027_88866463 | 0.17 |
Pim3 |
proviral integration site 3 |
1325 |
0.34 |
| chr7_98415563_98415739 | 0.17 |
Gm44507 |
predicted gene 44507 |
27 |
0.97 |
| chr10_98652216_98652384 | 0.17 |
Gm5427 |
predicted gene 5427 |
47410 |
0.18 |
| chr16_23967660_23967916 | 0.17 |
Bcl6 |
B cell leukemia/lymphoma 6 |
4592 |
0.19 |
| chr10_93492422_93492688 | 0.17 |
Hal |
histidine ammonia lyase |
3752 |
0.18 |
| chr9_121906088_121906239 | 0.16 |
Ackr2 |
atypical chemokine receptor 2 |
1 |
0.95 |
| chr11_97383324_97383535 | 0.16 |
Socs7 |
suppressor of cytokine signaling 7 |
5992 |
0.15 |
| chr11_117322484_117322692 | 0.16 |
Septin9 |
septin 9 |
3845 |
0.24 |
| chr14_41120981_41121358 | 0.16 |
Sftpa1 |
surfactant associated protein A1 |
10613 |
0.11 |
| chr17_86208576_86208727 | 0.16 |
AC154542.1 |
TEC |
33076 |
0.17 |
| chr11_101372782_101373038 | 0.16 |
G6pc |
glucose-6-phosphatase, catalytic |
5349 |
0.06 |
| chr6_34412803_34412954 | 0.16 |
Akr1b7 |
aldo-keto reductase family 1, member B7 |
544 |
0.69 |
| chr11_116180328_116180538 | 0.16 |
Acox1 |
acyl-Coenzyme A oxidase 1, palmitoyl |
781 |
0.44 |
| chr17_56005828_56005982 | 0.16 |
Mpnd |
MPN domain containing |
194 |
0.61 |
| chr17_8974900_8975051 | 0.16 |
1700010I14Rik |
RIKEN cDNA 1700010I14 gene |
13358 |
0.23 |
| chr12_110203702_110203885 | 0.16 |
Gm34785 |
predicted gene, 34785 |
15732 |
0.12 |
| chr6_86665663_86665814 | 0.15 |
Gm44292 |
predicted gene, 44292 |
187 |
0.89 |
| chr2_144308533_144308857 | 0.15 |
Ovol2 |
ovo like zinc finger 2 |
22631 |
0.09 |
| chr19_46029534_46029692 | 0.15 |
Ldb1 |
LIM domain binding 1 |
5265 |
0.14 |
| chr7_92681661_92681812 | 0.15 |
Pcf11 |
PCF11 cleavage and polyadenylation factor subunit |
11802 |
0.12 |
| chr2_155063372_155063735 | 0.15 |
Gm45609 |
predicted gene 45609 |
10628 |
0.13 |
| chr1_88960128_88960297 | 0.15 |
Gm4753 |
predicted gene 4753 |
27893 |
0.17 |
| chr5_120480072_120480223 | 0.15 |
Sds |
serine dehydratase |
216 |
0.8 |
| chr18_11144853_11145048 | 0.15 |
Gata6 |
GATA binding protein 6 |
85903 |
0.09 |
| chr7_84596857_84597020 | 0.15 |
Fah |
fumarylacetoacetate hydrolase |
5162 |
0.17 |
| chr4_41637175_41637517 | 0.15 |
Gm12406 |
predicted gene 12406 |
2032 |
0.17 |
| chr19_44997151_44997302 | 0.15 |
Sema4g |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
5090 |
0.11 |
| chr17_39846008_39846161 | 0.15 |
CT010467.1 |
18s RNA, related sequence 5 |
269 |
0.9 |
| chr6_134078837_134079020 | 0.15 |
Gm43982 |
predicted gene, 43982 |
5574 |
0.2 |
| chr3_102187285_102187460 | 0.15 |
Vangl1 |
VANGL planar cell polarity 1 |
10533 |
0.14 |
| chr4_150592192_150592343 | 0.14 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
22610 |
0.17 |
| chr9_78586353_78586504 | 0.14 |
Slc17a5 |
solute carrier family 17 (anion/sugar transporter), member 5 |
1587 |
0.33 |
| chr10_24911578_24911812 | 0.14 |
Mir6905 |
microRNA 6905 |
1031 |
0.42 |
| chr7_19496130_19496424 | 0.14 |
Exoc3l2 |
exocyst complex component 3-like 2 |
7221 |
0.08 |
| chr4_135231252_135231420 | 0.14 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
41478 |
0.11 |
| chr7_140765024_140765175 | 0.14 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
618 |
0.54 |
| chr11_98772948_98773272 | 0.14 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2223 |
0.16 |
| chr2_31473064_31473684 | 0.14 |
Ass1 |
argininosuccinate synthetase 1 |
3167 |
0.24 |
| chr14_101854215_101854466 | 0.14 |
Lmo7 |
LIM domain only 7 |
13521 |
0.26 |
| chr7_122971788_122971939 | 0.14 |
Rbbp6 |
retinoblastoma binding protein 6, ubiquitin ligase |
945 |
0.45 |
| chr11_115246492_115246668 | 0.14 |
Gm25837 |
predicted gene, 25837 |
6698 |
0.11 |
| chr17_9663030_9663198 | 0.14 |
Pabpc6 |
poly(A) binding protein, cytoplasmic 6 |
6603 |
0.26 |
| chr15_81481395_81481613 | 0.14 |
Gm49406 |
predicted gene, 49406 |
4877 |
0.14 |
| chr3_96567003_96567180 | 0.14 |
Gm15441 |
predicted gene 15441 |
290 |
0.76 |
| chr4_117125610_117126073 | 0.14 |
Btbd19 |
BTB (POZ) domain containing 19 |
116 |
0.86 |
| chr7_113913919_113914070 | 0.14 |
Spon1 |
spondin 1, (f-spondin) extracellular matrix protein |
147820 |
0.04 |
| chr4_152165673_152165840 | 0.14 |
Hes2 |
hes family bHLH transcription factor 2 |
6207 |
0.12 |
| chr17_46131243_46131394 | 0.14 |
Rsph9 |
radial spoke head 9 homolog (Chlamydomonas) |
5477 |
0.12 |
| chr7_113935146_113935310 | 0.14 |
Gm45615 |
predicted gene 45615 |
151670 |
0.04 |
| chr16_87690191_87690394 | 0.14 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
8653 |
0.22 |
| chr8_77486943_77487275 | 0.13 |
0610038B21Rik |
RIKEN cDNA 0610038B21 gene |
29947 |
0.14 |
| chr2_174343758_174344168 | 0.13 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
349 |
0.83 |
| chr8_36291164_36291366 | 0.13 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
41749 |
0.14 |
| chr15_31259637_31259788 | 0.13 |
Dap |
death-associated protein |
8624 |
0.16 |
| chr11_43889353_43889625 | 0.13 |
Adra1b |
adrenergic receptor, alpha 1b |
11721 |
0.25 |
| chr2_167722273_167722608 | 0.13 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
31259 |
0.1 |
| chr13_9052712_9052890 | 0.13 |
Gm36264 |
predicted gene, 36264 |
23650 |
0.12 |
| chr17_78714751_78714902 | 0.13 |
Strn |
striatin, calmodulin binding protein |
22370 |
0.14 |
| chr3_121195173_121195348 | 0.13 |
Gm5710 |
predicted gene 5710 |
14163 |
0.14 |
| chr19_46108526_46108716 | 0.13 |
Gm50306 |
predicted gene, 50306 |
16309 |
0.11 |
| chr6_48492947_48493147 | 0.13 |
Sspo |
SCO-spondin |
733 |
0.46 |
| chr4_127034401_127034573 | 0.13 |
Gm12940 |
predicted gene 12940 |
756 |
0.49 |
| chr4_109406664_109406986 | 0.13 |
Ttc39a |
tetratricopeptide repeat domain 39A |
202 |
0.83 |
| chr4_118149267_118149447 | 0.13 |
Kdm4a |
lysine (K)-specific demethylase 4A |
2598 |
0.22 |
| chr17_25756148_25756299 | 0.13 |
Msln |
mesothelin |
1845 |
0.11 |
| chr2_135854498_135854679 | 0.13 |
Plcb4 |
phospholipase C, beta 4 |
33106 |
0.19 |
| chr2_124902813_124902964 | 0.13 |
Slc24a5 |
solute carrier family 24, member 5 |
165236 |
0.03 |
| chr9_44665366_44665550 | 0.13 |
Gm3953 |
predicted gene 3953 |
742 |
0.4 |
| chr2_51823121_51823300 | 0.13 |
Gm13492 |
predicted gene 13492 |
4641 |
0.25 |
| chr1_186261473_186261624 | 0.13 |
Gm37272 |
predicted gene, 37272 |
79291 |
0.09 |
| chr15_97682260_97682559 | 0.13 |
Rpap3 |
RNA polymerase II associated protein 3 |
210 |
0.94 |
| chr5_33901574_33901762 | 0.13 |
Mir7024 |
microRNA 7024 |
2697 |
0.18 |
| chr11_102755530_102755686 | 0.13 |
Adam11 |
a disintegrin and metallopeptidase domain 11 |
5831 |
0.12 |
| chr6_72360016_72360452 | 0.13 |
Rnf181 |
ring finger protein 181 |
833 |
0.41 |
| chr17_34733622_34734232 | 0.13 |
C4b |
complement component 4B (Chido blood group) |
2172 |
0.12 |
| chr16_30755886_30756050 | 0.13 |
Gm49754 |
predicted gene, 49754 |
37287 |
0.15 |
| chr9_111148401_111148693 | 0.13 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
12756 |
0.17 |
| chr10_37247416_37247567 | 0.13 |
4930543K20Rik |
RIKEN cDNA 4930543K20 gene |
16257 |
0.24 |
| chr15_81355699_81355864 | 0.13 |
Slc25a17 |
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
4946 |
0.16 |
| chr8_126801964_126802115 | 0.13 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
37194 |
0.15 |
| chr15_81480824_81481310 | 0.13 |
Gm49406 |
predicted gene, 49406 |
5314 |
0.14 |
| chr6_121159588_121159765 | 0.13 |
Pex26 |
peroxisomal biogenesis factor 26 |
23991 |
0.12 |
| chr3_144109828_144110050 | 0.12 |
Gm34078 |
predicted gene, 34078 |
25815 |
0.2 |
| chr13_23285684_23285835 | 0.12 |
4933404K08Rik |
RIKEN cDNA 4933404K08 gene |
7409 |
0.08 |
| chr3_51252801_51253044 | 0.12 |
Elf2 |
E74-like factor 2 |
7319 |
0.13 |
| chr15_85354161_85354332 | 0.12 |
Gm23517 |
predicted gene, 23517 |
2992 |
0.27 |
| chr2_173633527_173633866 | 0.12 |
Ppp4r1l-ps |
protein phosphatase 4, regulatory subunit 1-like, pseudogene |
4146 |
0.21 |
| chr10_85103179_85103555 | 0.12 |
Tmem263 |
transmembrane protein 263 |
624 |
0.52 |
| chr6_138128655_138128834 | 0.12 |
Mgst1 |
microsomal glutathione S-transferase 1 |
11572 |
0.26 |
| chr16_45377470_45377621 | 0.12 |
Cd200 |
CD200 antigen |
22767 |
0.16 |
| chr7_45150438_45150589 | 0.12 |
Aldh16a1 |
aldehyde dehydrogenase 16 family, member A1 |
1334 |
0.13 |
| chr14_68390472_68390641 | 0.12 |
Gm31227 |
predicted gene, 31227 |
71075 |
0.11 |
| chr17_56074297_56074510 | 0.12 |
Ubxn6 |
UBX domain protein 6 |
144 |
0.9 |
| chr10_69126441_69126619 | 0.12 |
Gm24172 |
predicted gene, 24172 |
6352 |
0.17 |
| chr19_8952481_8952632 | 0.12 |
Tut1 |
terminal uridylyl transferase 1, U6 snRNA-specific |
1291 |
0.2 |
| chr13_80885948_80886129 | 0.12 |
Arrdc3 |
arrestin domain containing 3 |
57 |
0.89 |
| chr19_46587795_46587964 | 0.12 |
Sfxn2 |
sideroflexin 2 |
3242 |
0.18 |
| chr8_60491849_60492091 | 0.12 |
Gm10283 |
predicted gene 10283 |
11228 |
0.22 |
| chr1_67189193_67189422 | 0.12 |
Gm15668 |
predicted gene 15668 |
59893 |
0.11 |
| chr1_67134415_67135075 | 0.12 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
11719 |
0.24 |
| chr4_102966036_102966207 | 0.12 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
3765 |
0.19 |
| chr1_39192284_39192607 | 0.12 |
Npas2 |
neuronal PAS domain protein 2 |
1286 |
0.45 |
| chr11_119105318_119105491 | 0.12 |
Gm11754 |
predicted gene 11754 |
6021 |
0.15 |
| chr18_35277643_35277851 | 0.12 |
Gm50146 |
predicted gene, 50146 |
13718 |
0.16 |
| chr1_192061757_192061908 | 0.12 |
Traf5 |
TNF receptor-associated factor 5 |
2670 |
0.19 |
| chr2_164161443_164161616 | 0.12 |
n-R5s207 |
nuclear encoded rRNA 5S 207 |
4008 |
0.11 |
| chr7_145044142_145044344 | 0.12 |
Ccnd1 |
cyclin D1 |
104318 |
0.05 |
| chr11_102749526_102749741 | 0.12 |
Adam11 |
a disintegrin and metallopeptidase domain 11 |
11806 |
0.11 |
| chr14_76623735_76623902 | 0.12 |
Fkbp1a-ps1 |
FK506 binding protein 1a, pseudogene 1 |
13046 |
0.21 |
| chr4_118247695_118247892 | 0.12 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
11348 |
0.17 |
| chr11_116389158_116389441 | 0.12 |
Rnf157 |
ring finger protein 157 |
1894 |
0.21 |
| chr10_71857688_71857856 | 0.12 |
Gm22937 |
predicted gene, 22937 |
40264 |
0.15 |
| chr3_130660210_130660361 | 0.12 |
Etnppl |
ethanolamine phosphate phospholyase |
33765 |
0.11 |
| chr2_31496143_31496706 | 0.12 |
Ass1 |
argininosuccinate synthetase 1 |
1348 |
0.43 |
| chr12_75851882_75852228 | 0.12 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
675 |
0.77 |
| chr8_36223220_36223376 | 0.12 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
26218 |
0.14 |
| chr11_3287201_3287367 | 0.12 |
Patz1 |
POZ (BTB) and AT hook containing zinc finger 1 |
1590 |
0.26 |
| chr14_30909356_30909712 | 0.11 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
4595 |
0.13 |
| chr19_10187571_10187722 | 0.11 |
Mir6993 |
microRNA 6993 |
3729 |
0.12 |
| chr13_111601521_111601690 | 0.11 |
Gm10736 |
predicted gene 10736 |
7994 |
0.13 |
| chr8_71377251_71377402 | 0.11 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
1536 |
0.21 |
| chr17_56273355_56273537 | 0.11 |
Ticam1 |
toll-like receptor adaptor molecule 1 |
3340 |
0.12 |
| chr10_7852226_7852377 | 0.11 |
Gm24726 |
predicted gene, 24726 |
3820 |
0.16 |
| chr4_95640632_95640805 | 0.11 |
Fggy |
FGGY carbohydrate kinase domain containing |
3831 |
0.31 |
| chr8_106074831_106075102 | 0.11 |
Nfatc3 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
15333 |
0.1 |
| chr6_120926829_120927234 | 0.11 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
8504 |
0.12 |
| chr5_28076176_28076405 | 0.11 |
Insig1 |
insulin induced gene 1 |
1624 |
0.34 |
| chr5_107874706_107874894 | 0.11 |
Evi5 |
ecotropic viral integration site 5 |
244 |
0.86 |
| chr8_108745611_108745766 | 0.11 |
Gm38042 |
predicted gene, 38042 |
8095 |
0.24 |
| chr18_53927118_53927269 | 0.11 |
Csnk1g3 |
casein kinase 1, gamma 3 |
5051 |
0.29 |
| chr8_116351879_116352235 | 0.11 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
5004 |
0.32 |
| chr9_67869712_67870477 | 0.11 |
Vps13c |
vacuolar protein sorting 13C |
29682 |
0.17 |
| chr7_113228562_113228716 | 0.11 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
5984 |
0.23 |
| chr15_99096799_99097225 | 0.11 |
Dnajc22 |
DnaJ heat shock protein family (Hsp40) member C22 |
2400 |
0.14 |
| chr1_98713946_98714114 | 0.11 |
Gm29460 |
predicted gene 29460 |
50811 |
0.17 |
| chr11_98459290_98459441 | 0.11 |
Grb7 |
growth factor receptor bound protein 7 |
5576 |
0.1 |
| chr8_124573046_124573209 | 0.11 |
Capn9 |
calpain 9 |
2984 |
0.21 |
| chrX_12189621_12189798 | 0.11 |
Gm14521 |
predicted gene 14521 |
9642 |
0.24 |
| chr14_65972599_65972894 | 0.11 |
Clu |
clusterin |
1443 |
0.35 |
| chr17_31319478_31319804 | 0.11 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
465 |
0.76 |
| chr7_99935891_99936042 | 0.11 |
Gm18959 |
predicted gene, 18959 |
9096 |
0.12 |
| chr16_4371080_4371246 | 0.11 |
Adcy9 |
adenylate cyclase 9 |
48424 |
0.12 |
| chr15_12010383_12010678 | 0.11 |
Sub1 |
SUB1 homolog, transcriptional regulator |
13547 |
0.16 |
| chr10_81408031_81408226 | 0.11 |
Nfic |
nuclear factor I/C |
487 |
0.49 |
| chr7_63922331_63923024 | 0.11 |
Klf13 |
Kruppel-like factor 13 |
2193 |
0.22 |
| chr15_75827912_75828063 | 0.11 |
Zc3h3 |
zinc finger CCCH type containing 3 |
222 |
0.88 |
| chr1_188813741_188813916 | 0.11 |
Gm25269 |
predicted gene, 25269 |
53352 |
0.16 |
| chr19_16181317_16181503 | 0.11 |
E030024N20Rik |
RIKEN cDNA E030024N20 gene |
16605 |
0.2 |
| chr15_31252945_31253125 | 0.11 |
Dap |
death-associated protein |
15301 |
0.15 |
| chr5_53278040_53278387 | 0.11 |
Smim20 |
small integral membrane protein 20 |
1095 |
0.48 |
| chr6_72158619_72158820 | 0.11 |
Gm38832 |
predicted gene, 38832 |
4130 |
0.17 |
| chr2_31485093_31485244 | 0.11 |
Ass1 |
argininosuccinate synthetase 1 |
12604 |
0.18 |
| chr4_104822600_104822778 | 0.11 |
C8b |
complement component 8, beta polypeptide |
23481 |
0.18 |
| chr2_167959698_167959868 | 0.11 |
Ptpn1 |
protein tyrosine phosphatase, non-receptor type 1 |
8735 |
0.17 |
| chr9_116228629_116228809 | 0.11 |
Gm31410 |
predicted gene, 31410 |
9917 |
0.19 |
| chr7_24316221_24316527 | 0.11 |
Zfp94 |
zinc finger protein 94 |
208 |
0.82 |
| chr7_44589314_44589478 | 0.11 |
Gm15396 |
predicted gene 15396 |
1228 |
0.19 |
| chr15_90679035_90679205 | 0.11 |
Cpne8 |
copine VIII |
251 |
0.93 |
| chr1_181465127_181465294 | 0.11 |
Gm16539 |
predicted gene 16539 |
32810 |
0.14 |
| chr2_31505484_31505674 | 0.11 |
Ass1 |
argininosuccinate synthetase 1 |
4853 |
0.2 |
| chr11_109502246_109502399 | 0.11 |
Gm22378 |
predicted gene, 22378 |
2498 |
0.21 |
| chr13_93640508_93640869 | 0.11 |
Bhmt |
betaine-homocysteine methyltransferase |
2727 |
0.21 |
| chr10_69190805_69191022 | 0.10 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
17639 |
0.16 |
| chr2_117018527_117018695 | 0.10 |
Gm13981 |
predicted gene 13981 |
36955 |
0.14 |
| chr2_59979903_59980091 | 0.10 |
Gm13574 |
predicted gene 13574 |
12347 |
0.19 |
| chr10_42205235_42205454 | 0.10 |
Foxo3 |
forkhead box O3 |
53022 |
0.15 |
| chr14_25741793_25741944 | 0.10 |
Zcchc24 |
zinc finger, CCHC domain containing 24 |
22076 |
0.13 |
| chr4_118476510_118476661 | 0.10 |
Tie1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
13170 |
0.1 |
| chr19_47305501_47305661 | 0.10 |
Sh3pxd2a |
SH3 and PX domains 2A |
9170 |
0.17 |
| chr15_78852145_78852296 | 0.10 |
Lgals2 |
lectin, galactose-binding, soluble 2 |
3309 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.0 | 0.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.0 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0043731 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |