Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
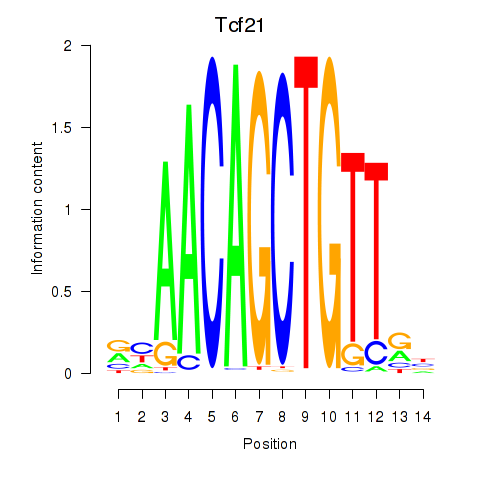
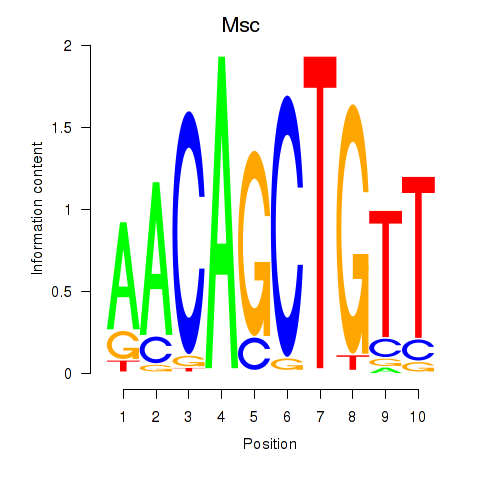
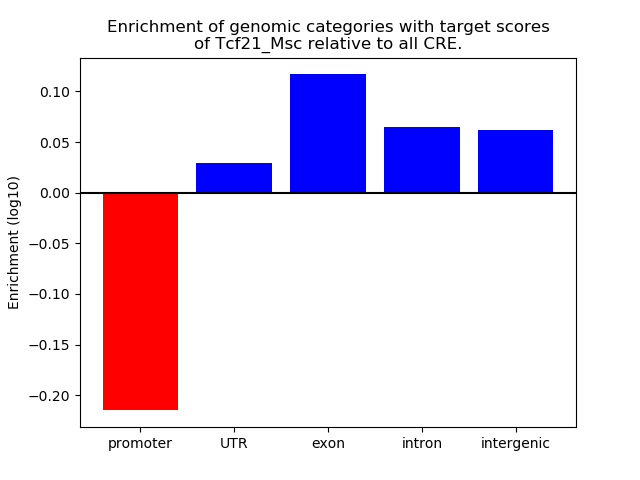
Results for Tcf21_Msc
Z-value: 1.61
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf21
|
ENSMUSG00000045680.7 | transcription factor 21 |
|
Msc
|
ENSMUSG00000025930.5 | musculin |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_22820121_22820272 | Tcf21 | 22 | 0.972710 | 0.26 | 6.1e-01 | Click! |
| chr10_22843285_22843469 | Tcf21 | 23203 | 0.157237 | -0.07 | 8.9e-01 | Click! |
| chr10_22875325_22875476 | Tcf21 | 55226 | 0.108644 | -0.02 | 9.7e-01 | Click! |
Activity of the Tcf21_Msc motif across conditions
Conditions sorted by the z-value of the Tcf21_Msc motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_121802332_121802546 | 1.11 |
Gm44740 |
predicted gene 44740 |
36626 |
0.13 |
| chr14_30173289_30173526 | 1.10 |
Cacna1d |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
1984 |
0.35 |
| chr13_58962693_58962856 | 0.87 |
Ntrk2 |
neurotrophic tyrosine kinase, receptor, type 2 |
91039 |
0.06 |
| chr5_30534594_30534940 | 0.84 |
Gm42765 |
predicted gene 42765 |
5267 |
0.14 |
| chr9_74893395_74893777 | 0.80 |
Onecut1 |
one cut domain, family member 1 |
27102 |
0.13 |
| chr11_6021664_6021840 | 0.72 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
21304 |
0.15 |
| chr19_10599348_10599499 | 0.72 |
Tkfc |
triokinase, FMN cyclase |
4835 |
0.1 |
| chr9_74884431_74884582 | 0.70 |
Onecut1 |
one cut domain, family member 1 |
18022 |
0.15 |
| chr6_149333094_149333245 | 0.68 |
Resf1 |
retroelement silencing factor 1 |
7711 |
0.14 |
| chr10_95434881_95435051 | 0.68 |
5730420D15Rik |
RIKEN cDNA 5730420D15 gene |
17591 |
0.11 |
| chr18_84376797_84376986 | 0.66 |
Gm37216 |
predicted gene, 37216 |
745 |
0.73 |
| chr6_93669779_93669930 | 0.63 |
Gm7812 |
predicted gene 7812 |
19629 |
0.25 |
| chr11_16821774_16821942 | 0.63 |
Egfros |
epidermal growth factor receptor, opposite strand |
8844 |
0.22 |
| chr15_39116967_39117141 | 0.60 |
Dcaf13 |
DDB1 and CUL4 associated factor 13 |
4189 |
0.15 |
| chr5_43310353_43310569 | 0.59 |
Gm43020 |
predicted gene 43020 |
44798 |
0.11 |
| chr8_4839167_4839331 | 0.59 |
Gm44961 |
predicted gene 44961 |
18019 |
0.12 |
| chr8_3211565_3211796 | 0.57 |
Insr |
insulin receptor |
18963 |
0.18 |
| chr2_90668038_90668212 | 0.57 |
Nup160 |
nucleoporin 160 |
9090 |
0.2 |
| chr11_76827089_76827249 | 0.56 |
Cpd |
carboxypeptidase D |
18343 |
0.19 |
| chr11_16879295_16879446 | 0.56 |
Egfr |
epidermal growth factor receptor |
1220 |
0.5 |
| chr13_80890166_80890317 | 0.56 |
Arrdc3 |
arrestin domain containing 3 |
137 |
0.95 |
| chr11_120817967_120818292 | 0.56 |
Fasn |
fatty acid synthase |
2674 |
0.13 |
| chr3_152091714_152091865 | 0.53 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
11065 |
0.14 |
| chr11_16879568_16879719 | 0.53 |
Egfr |
epidermal growth factor receptor |
1493 |
0.43 |
| chr8_46385009_46385281 | 0.53 |
Gm45253 |
predicted gene 45253 |
743 |
0.6 |
| chr12_104087488_104087669 | 0.53 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
6929 |
0.1 |
| chr11_120814981_120815132 | 0.52 |
Fasn |
fatty acid synthase |
399 |
0.71 |
| chr2_167292312_167292496 | 0.52 |
Gm11473 |
predicted gene 11473 |
10627 |
0.16 |
| chr4_141955631_141955810 | 0.52 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
1526 |
0.3 |
| chr11_16879987_16880147 | 0.51 |
Egfr |
epidermal growth factor receptor |
1917 |
0.36 |
| chr4_117831574_117831756 | 0.51 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
2841 |
0.16 |
| chr8_120046709_120046884 | 0.51 |
Gm15684 |
predicted gene 15684 |
1509 |
0.32 |
| chr6_117577757_117577908 | 0.51 |
Gm9946 |
predicted gene 9946 |
10160 |
0.2 |
| chr9_116136919_116137123 | 0.51 |
Tgfbr2 |
transforming growth factor, beta receptor II |
38244 |
0.15 |
| chr11_16828832_16828991 | 0.49 |
Egfros |
epidermal growth factor receptor, opposite strand |
1791 |
0.4 |
| chrX_42602450_42602620 | 0.47 |
Sh2d1a |
SH2 domain containing 1A |
75950 |
0.1 |
| chr12_104084479_104084838 | 0.47 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
4009 |
0.12 |
| chr15_87233626_87233777 | 0.46 |
A930027H12Rik |
RIKEN cDNA A930027H12 gene |
9849 |
0.25 |
| chr11_52257389_52257568 | 0.45 |
Tcf7 |
transcription factor 7, T cell specific |
17399 |
0.11 |
| chr3_69362162_69362601 | 0.45 |
Gm17212 |
predicted gene 17212 |
26621 |
0.16 |
| chr1_131770748_131770959 | 0.45 |
Slc26a9 |
solute carrier family 26, member 9 |
20362 |
0.14 |
| chr14_9608900_9609103 | 0.45 |
Gm48371 |
predicted gene, 48371 |
139446 |
0.05 |
| chr2_156571655_156571819 | 0.44 |
Gm14168 |
predicted gene 14168 |
55 |
0.96 |
| chr14_32708910_32709089 | 0.44 |
Gm28651 |
predicted gene 28651 |
22441 |
0.16 |
| chr18_6464158_6464366 | 0.44 |
Epc1 |
enhancer of polycomb homolog 1 |
22368 |
0.15 |
| chr17_86446823_86447020 | 0.44 |
Prkce |
protein kinase C, epsilon |
43641 |
0.17 |
| chr10_111314954_111315105 | 0.44 |
Bbs10 |
Bardet-Biedl syndrome 10 (human) |
16350 |
0.18 |
| chr5_125524109_125525122 | 0.44 |
Tmem132b |
transmembrane protein 132B |
7159 |
0.16 |
| chr7_79308613_79308815 | 0.43 |
Gm39041 |
predicted gene, 39041 |
9689 |
0.14 |
| chr5_125492454_125492640 | 0.43 |
Gm27551 |
predicted gene, 27551 |
13170 |
0.13 |
| chr9_31610718_31610869 | 0.42 |
Gm18820 |
predicted gene, 18820 |
32977 |
0.14 |
| chr19_47660118_47660275 | 0.42 |
Col17a1 |
collagen, type XVII, alpha 1 |
10601 |
0.12 |
| chr11_120819185_120819682 | 0.42 |
Fasn |
fatty acid synthase |
3978 |
0.11 |
| chr19_11977402_11977711 | 0.41 |
Osbp |
oxysterol binding protein |
11615 |
0.09 |
| chr1_36316895_36317108 | 0.41 |
Arid5a |
AT rich interactive domain 5A (MRF1-like) |
26 |
0.97 |
| chr18_84376572_84376752 | 0.41 |
Gm37216 |
predicted gene, 37216 |
516 |
0.84 |
| chr17_70004735_70004886 | 0.41 |
Dlgap1 |
DLG associated protein 1 |
35389 |
0.2 |
| chr10_87880307_87880601 | 0.41 |
Igf1os |
insulin-like growth factor 1, opposite strand |
17073 |
0.18 |
| chr8_34460252_34460438 | 0.41 |
Gm45349 |
predicted gene 45349 |
12650 |
0.18 |
| chr4_10854377_10854556 | 0.40 |
Gm12919 |
predicted gene 12919 |
623 |
0.61 |
| chr1_138967552_138967703 | 0.40 |
Dennd1b |
DENN/MADD domain containing 1B |
93 |
0.91 |
| chr5_139358617_139359046 | 0.40 |
Cyp2w1 |
cytochrome P450, family 2, subfamily w, polypeptide 1 |
4255 |
0.13 |
| chr5_75063247_75063407 | 0.40 |
Gsx2 |
GS homeobox 2 |
12274 |
0.11 |
| chr16_56063021_56063172 | 0.39 |
Gm24047 |
predicted gene, 24047 |
7467 |
0.12 |
| chr9_74891211_74891440 | 0.39 |
Onecut1 |
one cut domain, family member 1 |
24841 |
0.14 |
| chr7_127973702_127973919 | 0.39 |
Fus |
fused in sarcoma |
333 |
0.72 |
| chr18_65989569_65989860 | 0.38 |
Lman1 |
lectin, mannose-binding, 1 |
3946 |
0.15 |
| chr13_112331307_112331468 | 0.38 |
Ankrd55 |
ankyrin repeat domain 55 |
12933 |
0.17 |
| chr12_41500503_41500654 | 0.37 |
Lrrn3 |
leucine rich repeat protein 3, neuronal |
14147 |
0.22 |
| chr11_120811259_120811452 | 0.37 |
Fasn |
fatty acid synthase |
1654 |
0.19 |
| chr18_61357667_61357827 | 0.37 |
Gm25301 |
predicted gene, 25301 |
40066 |
0.1 |
| chr18_65968667_65968818 | 0.37 |
Cplx4 |
complexin 4 |
1060 |
0.37 |
| chr9_55278709_55278860 | 0.36 |
Nrg4 |
neuregulin 4 |
4788 |
0.2 |
| chr7_15939282_15939433 | 0.36 |
Snord23 |
small nucleolar RNA, C/D box 23 |
487 |
0.65 |
| chr5_33465411_33465562 | 0.36 |
Gm43851 |
predicted gene 43851 |
28022 |
0.15 |
| chr5_125484345_125484496 | 0.36 |
Gm27551 |
predicted gene, 27551 |
5043 |
0.14 |
| chrX_111626357_111626522 | 0.36 |
Gm7340 |
predicted gene 7340 |
54740 |
0.14 |
| chr13_58192282_58192443 | 0.35 |
Gm48357 |
predicted gene, 48357 |
1044 |
0.37 |
| chr9_98418368_98418712 | 0.35 |
Rbp1 |
retinol binding protein 1, cellular |
4421 |
0.24 |
| chr8_105256295_105256446 | 0.35 |
B3gnt9 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
1217 |
0.21 |
| chr10_91261198_91261441 | 0.35 |
Gm18705 |
predicted gene, 18705 |
1975 |
0.31 |
| chrX_56970947_56971107 | 0.35 |
Adgrg4 |
adhesion G protein-coupled receptor G4 |
2907 |
0.31 |
| chr4_44784029_44784206 | 0.35 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
23673 |
0.15 |
| chr9_35103547_35103698 | 0.35 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
3045 |
0.2 |
| chr9_34258652_34258813 | 0.35 |
Gm27161 |
predicted gene 27161 |
58504 |
0.15 |
| chr5_125523118_125523269 | 0.35 |
Aacs |
acetoacetyl-CoA synthetase |
7950 |
0.16 |
| chr11_16808080_16808304 | 0.34 |
Egfros |
epidermal growth factor receptor, opposite strand |
22510 |
0.19 |
| chr5_144416866_144417038 | 0.34 |
Gm15709 |
predicted gene 15709 |
10350 |
0.2 |
| chr11_118451300_118451612 | 0.34 |
Gm11747 |
predicted gene 11747 |
3539 |
0.16 |
| chr11_86769997_86770170 | 0.34 |
Cltc |
clathrin, heavy polypeptide (Hc) |
12518 |
0.19 |
| chr13_48916144_48916308 | 0.34 |
Phf2 |
PHD finger protein 2 |
45107 |
0.13 |
| chr1_92683263_92683419 | 0.34 |
Gm29483 |
predicted gene 29483 |
33861 |
0.09 |
| chr10_80668723_80668916 | 0.33 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
3157 |
0.11 |
| chr11_60201760_60201955 | 0.33 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
358 |
0.51 |
| chr1_50806470_50806812 | 0.32 |
Gm28321 |
predicted gene 28321 |
8734 |
0.28 |
| chrX_77601507_77601673 | 0.32 |
Gm23121 |
predicted gene, 23121 |
22103 |
0.2 |
| chr17_10093572_10093736 | 0.32 |
Qk |
quaking |
119920 |
0.06 |
| chr8_80289362_80289553 | 0.32 |
Gm45430 |
predicted gene 45430 |
94258 |
0.08 |
| chr10_8818073_8818257 | 0.32 |
Gm25410 |
predicted gene, 25410 |
5982 |
0.21 |
| chr14_63326787_63326957 | 0.32 |
Gm5463 |
predicted gene 5463 |
2348 |
0.25 |
| chr7_38162494_38162645 | 0.32 |
Gm22203 |
predicted gene, 22203 |
14102 |
0.13 |
| chr4_150660019_150660333 | 0.32 |
Slc45a1 |
solute carrier family 45, member 1 |
8002 |
0.19 |
| chr17_43270317_43270505 | 0.32 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
82 |
0.98 |
| chr4_97765098_97765412 | 0.32 |
Nfia |
nuclear factor I/A |
7479 |
0.2 |
| chr12_30372401_30372579 | 0.32 |
Sntg2 |
syntrophin, gamma 2 |
775 |
0.73 |
| chr4_129216612_129216763 | 0.32 |
Yars |
tyrosyl-tRNA synthetase |
2925 |
0.17 |
| chr12_112125883_112126333 | 0.32 |
Mir203 |
microRNA 203 |
4772 |
0.13 |
| chr4_57070625_57070924 | 0.32 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
215 |
0.95 |
| chr11_16878966_16879155 | 0.32 |
Egfr |
epidermal growth factor receptor |
910 |
0.61 |
| chr10_117884880_117885031 | 0.31 |
Gm32552 |
predicted gene, 32552 |
18240 |
0.14 |
| chr8_105090981_105091315 | 0.31 |
Ces3b |
carboxylesterase 3B |
2529 |
0.16 |
| chr11_6158649_6158802 | 0.31 |
Rps15a-ps6 |
ribosomal protein S15A, pseudogene 6 |
14311 |
0.13 |
| chr1_174931061_174931246 | 0.30 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
9334 |
0.3 |
| chr6_67300933_67301090 | 0.30 |
Serbp1 |
serpine1 mRNA binding protein 1 |
29132 |
0.12 |
| chr1_151686797_151686981 | 0.30 |
Fam129a |
family with sequence similarity 129, member A |
9722 |
0.22 |
| chr5_149616281_149616432 | 0.30 |
Hsph1 |
heat shock 105kDa/110kDa protein 1 |
2817 |
0.21 |
| chr2_58768981_58769132 | 0.30 |
Upp2 |
uridine phosphorylase 2 |
3731 |
0.25 |
| chr2_84995821_84996178 | 0.30 |
Prg3 |
proteoglycan 3 |
7784 |
0.12 |
| chr6_119643983_119644181 | 0.30 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
21977 |
0.22 |
| chr15_59454105_59454285 | 0.30 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
79908 |
0.08 |
| chr2_58771897_58772048 | 0.30 |
Upp2 |
uridine phosphorylase 2 |
6647 |
0.22 |
| chr15_55634841_55635006 | 0.30 |
Mtbp |
Mdm2, transformed 3T3 cell double minute p53 binding protein |
13471 |
0.25 |
| chr11_98766772_98767040 | 0.30 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3520 |
0.12 |
| chr15_27800508_27800681 | 0.30 |
Trio |
triple functional domain (PTPRF interacting) |
11956 |
0.22 |
| chr15_3524128_3524292 | 0.30 |
Ghr |
growth hormone receptor |
52566 |
0.15 |
| chr2_166834206_166834387 | 0.30 |
Arfgef2 |
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
6639 |
0.19 |
| chr15_100112289_100112692 | 0.30 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
4329 |
0.17 |
| chr2_158317109_158317280 | 0.29 |
Lbp |
lipopolysaccharide binding protein |
2988 |
0.15 |
| chr13_43530948_43531160 | 0.29 |
Gm32939 |
predicted gene, 32939 |
834 |
0.53 |
| chr11_95370162_95370363 | 0.29 |
Fam117a |
family with sequence similarity 117, member A |
5205 |
0.13 |
| chr5_138863743_138864096 | 0.29 |
Gm5294 |
predicted gene 5294 |
43839 |
0.13 |
| chr18_38016662_38016813 | 0.29 |
Gm30093 |
predicted gene, 30093 |
6792 |
0.12 |
| chr16_30664084_30664254 | 0.29 |
Fam43a |
family with sequence similarity 43, member A |
64446 |
0.1 |
| chr4_133561732_133562055 | 0.29 |
Gm23158 |
predicted gene, 23158 |
2944 |
0.13 |
| chr11_113667224_113667404 | 0.29 |
Cog1 |
component of oligomeric golgi complex 1 |
8031 |
0.12 |
| chr15_81249111_81249532 | 0.29 |
8430426J06Rik |
RIKEN cDNA 8430426J06 gene |
1353 |
0.36 |
| chr13_47039268_47039419 | 0.29 |
Tpmt |
thiopurine methyltransferase |
2238 |
0.17 |
| chr2_131869043_131869244 | 0.28 |
Erv3 |
endogenous retroviral sequence 3 |
9396 |
0.12 |
| chr9_83951310_83951505 | 0.28 |
Bckdhb |
branched chain ketoacid dehydrogenase E1, beta polypeptide |
2626 |
0.23 |
| chr1_90161712_90161886 | 0.28 |
4930434B07Rik |
RIKEN cDNA 4930434B07 gene |
8171 |
0.18 |
| chr6_90592647_90592924 | 0.28 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
3435 |
0.17 |
| chr8_84351080_84351276 | 0.28 |
Cacna1a |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
12539 |
0.13 |
| chr11_16771754_16772101 | 0.28 |
Egfr |
epidermal growth factor receptor |
19697 |
0.18 |
| chr1_162892140_162892544 | 0.28 |
Fmo2 |
flavin containing monooxygenase 2 |
5827 |
0.19 |
| chr10_68102616_68102812 | 0.28 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
33912 |
0.17 |
| chr1_153699524_153699675 | 0.28 |
Gm29529 |
predicted gene 29529 |
6943 |
0.12 |
| chr4_150080461_150080620 | 0.28 |
Gpr157 |
G protein-coupled receptor 157 |
6825 |
0.11 |
| chr9_74793041_74793363 | 0.28 |
Gm22315 |
predicted gene, 22315 |
11132 |
0.18 |
| chr3_18482157_18482333 | 0.28 |
4930433B08Rik |
RIKEN cDNA 4930433B08 gene |
1768 |
0.37 |
| chr13_38318516_38318698 | 0.27 |
Gm47990 |
predicted gene, 47990 |
1919 |
0.29 |
| chr5_102011072_102011265 | 0.27 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
29610 |
0.16 |
| chr7_115855107_115855272 | 0.27 |
Sox6 |
SRY (sex determining region Y)-box 6 |
4663 |
0.32 |
| chr11_106412764_106412932 | 0.27 |
Icam2 |
intercellular adhesion molecule 2 |
24773 |
0.12 |
| chr15_59319862_59320035 | 0.27 |
Sqle |
squalene epoxidase |
4841 |
0.18 |
| chr15_59004329_59004523 | 0.27 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
20290 |
0.16 |
| chr15_98738867_98739043 | 0.27 |
Fkbp11 |
FK506 binding protein 11 |
10757 |
0.09 |
| chr7_67636165_67636335 | 0.27 |
Gm45100 |
predicted gene 45100 |
1515 |
0.31 |
| chr1_41557400_41557596 | 0.27 |
Gm28634 |
predicted gene 28634 |
27955 |
0.26 |
| chr1_87763908_87764120 | 0.27 |
Atg16l1 |
autophagy related 16-like 1 (S. cerevisiae) |
2887 |
0.18 |
| chr12_32707351_32707528 | 0.27 |
Gm18726 |
predicted gene, 18726 |
2722 |
0.31 |
| chr11_118748971_118749122 | 0.27 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
11995 |
0.22 |
| chr14_74924136_74924288 | 0.27 |
Gm10847 |
predicted gene 10847 |
2233 |
0.32 |
| chr8_114128875_114129050 | 0.27 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
4595 |
0.32 |
| chr12_103687063_103687254 | 0.27 |
Serpina1f |
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
7508 |
0.1 |
| chr11_107850492_107850643 | 0.27 |
Gm27595 |
predicted gene, 27595 |
1178 |
0.49 |
| chr19_41482156_41482948 | 0.27 |
Lcor |
ligand dependent nuclear receptor corepressor |
85 |
0.98 |
| chr5_51144711_51144875 | 0.27 |
Gm44377 |
predicted gene, 44377 |
78881 |
0.1 |
| chr8_25256824_25256983 | 0.27 |
Tacc1 |
transforming, acidic coiled-coil containing protein 1 |
315 |
0.9 |
| chr19_20723027_20723214 | 0.26 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
4442 |
0.3 |
| chr16_32698159_32698320 | 0.26 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
17425 |
0.13 |
| chr9_108401502_108401859 | 0.26 |
1700102P08Rik |
RIKEN cDNA 1700102P08 gene |
8846 |
0.07 |
| chr7_26309085_26309252 | 0.26 |
Gm42375 |
predicted gene, 42375 |
1882 |
0.22 |
| chr11_116057344_116057497 | 0.26 |
Unk |
unkempt family zinc finger |
1522 |
0.24 |
| chr10_67696602_67696753 | 0.26 |
Gm47574 |
predicted gene, 47574 |
11161 |
0.16 |
| chr8_123661291_123661442 | 0.26 |
Rhou |
ras homolog family member U |
7437 |
0.05 |
| chrX_164521600_164521775 | 0.26 |
Asb9 |
ankyrin repeat and SOCS box-containing 9 |
15360 |
0.22 |
| chr2_173682841_173683018 | 0.26 |
Mir6340 |
microRNA 6340 |
18219 |
0.13 |
| chrX_36766114_36766265 | 0.26 |
Gm23613 |
predicted gene, 23613 |
10430 |
0.14 |
| chr11_84205927_84206078 | 0.26 |
Acaca |
acetyl-Coenzyme A carboxylase alpha |
10564 |
0.18 |
| chr1_41551537_41551723 | 0.26 |
Gm28634 |
predicted gene 28634 |
22087 |
0.28 |
| chr17_35784068_35784237 | 0.26 |
4833427F10Rik |
RIKEN cDNA 4833427F10 gene |
11702 |
0.08 |
| chr10_93431595_93431770 | 0.26 |
Mir7688 |
microRNA 7688 |
1425 |
0.36 |
| chr15_58983651_58983950 | 0.26 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
336 |
0.87 |
| chr18_12743258_12743476 | 0.26 |
Gm7599 |
predicted gene 7599 |
108 |
0.94 |
| chr12_79905913_79906084 | 0.25 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
18735 |
0.2 |
| chr3_10370926_10371217 | 0.25 |
Chmp4c |
charged multivesicular body protein 4C |
4113 |
0.15 |
| chr5_137052026_137052177 | 0.25 |
Ap1s1 |
adaptor protein complex AP-1, sigma 1 |
5966 |
0.11 |
| chr3_11357506_11357684 | 0.25 |
Gm22547 |
predicted gene, 22547 |
203258 |
0.03 |
| chr3_18105527_18105678 | 0.25 |
Gm23726 |
predicted gene, 23726 |
36276 |
0.16 |
| chr5_33720967_33721166 | 0.25 |
Fgfr3 |
fibroblast growth factor receptor 3 |
608 |
0.58 |
| chr4_140283535_140283688 | 0.25 |
Igsf21 |
immunoglobulin superfamily, member 21 |
36827 |
0.14 |
| chr14_67917259_67917426 | 0.25 |
Dock5 |
dedicator of cytokinesis 5 |
16090 |
0.22 |
| chr15_81798335_81798552 | 0.25 |
Tef |
thyrotroph embryonic factor |
3978 |
0.12 |
| chr5_97079576_97079732 | 0.25 |
Paqr3 |
progestin and adipoQ receptor family member III |
18117 |
0.16 |
| chr13_44306877_44307103 | 0.25 |
Gm29676 |
predicted gene, 29676 |
26541 |
0.17 |
| chr12_69813103_69813302 | 0.25 |
Map4k5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
5251 |
0.14 |
| chr4_139454119_139454298 | 0.25 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
9051 |
0.18 |
| chr2_152359606_152359999 | 0.25 |
Gm14165 |
predicted gene 14165 |
6187 |
0.1 |
| chr1_193928644_193928965 | 0.25 |
Gm21362 |
predicted gene, 21362 |
61795 |
0.15 |
| chr3_115815549_115815774 | 0.25 |
Dph5 |
diphthamide biosynthesis 5 |
72176 |
0.07 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.7 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.4 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.4 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.2 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.0 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.2 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.0 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.0 | GO:0045764 | positive regulation of cellular amino acid metabolic process(GO:0045764) |
| 0.0 | 0.0 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.0 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.0 | GO:0033121 | regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:1901723 | negative regulation of cell proliferation involved in kidney development(GO:1901723) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.0 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.3 | GO:2000758 | positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.2 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.0 | GO:0051610 | serotonin uptake(GO:0051610) |
| 0.0 | 0.0 | GO:1900125 | regulation of hyaluronan biosynthetic process(GO:1900125) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:1904752 | vascular associated smooth muscle cell migration(GO:1904738) regulation of vascular associated smooth muscle cell migration(GO:1904752) |
| 0.0 | 0.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:0060544 | regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0046719 | regulation by virus of viral protein levels in host cell(GO:0046719) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.0 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.2 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.8 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0030351 | inositol-1,3,4,5,6-pentakisphosphate 3-phosphatase activity(GO:0030351) inositol-1,4,5,6-tetrakisphosphate 6-phosphatase activity(GO:0030352) inositol tetrakisphosphate phosphatase activity(GO:0052743) inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |