Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
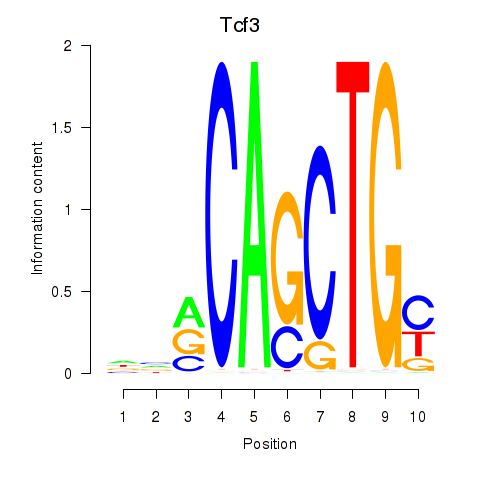
Results for Tcf3
Z-value: 1.36
Transcription factors associated with Tcf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf3
|
ENSMUSG00000020167.8 | transcription factor 3 |
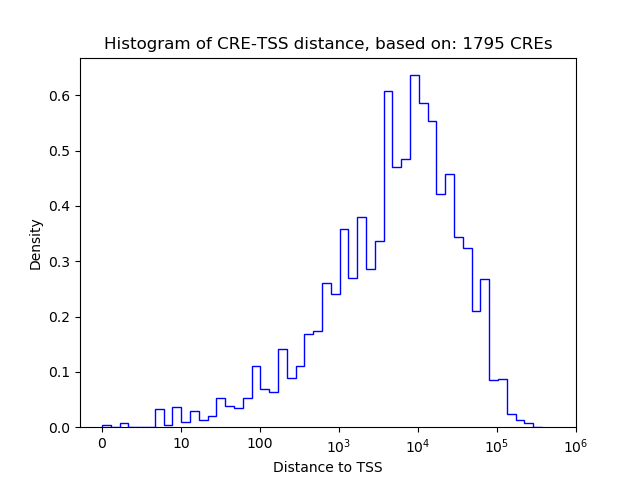
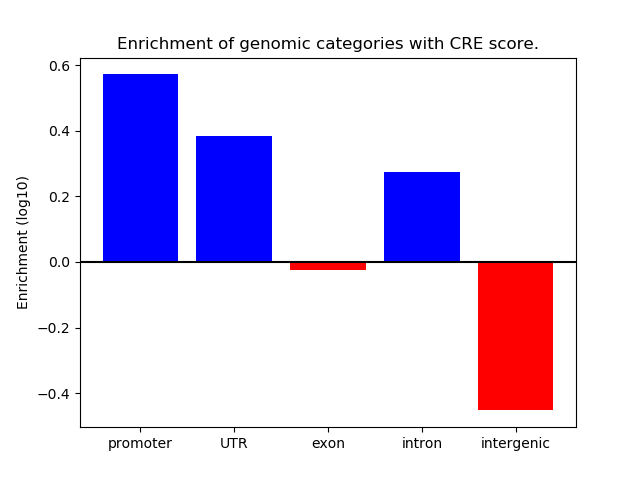
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_80423960_80424130 | Tcf3 | 2185 | 0.140379 | -0.98 | 6.2e-04 | Click! |
| chr10_80444503_80444654 | Tcf3 | 10931 | 0.085381 | -0.74 | 9.3e-02 | Click! |
| chr10_80433536_80433845 | Tcf3 | 43 | 0.943195 | -0.69 | 1.3e-01 | Click! |
| chr10_80446179_80446497 | Tcf3 | 12691 | 0.083839 | -0.66 | 1.5e-01 | Click! |
| chr10_80435895_80436307 | Tcf3 | 2454 | 0.136501 | -0.64 | 1.7e-01 | Click! |
Activity of the Tcf3 motif across conditions
Conditions sorted by the z-value of the Tcf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
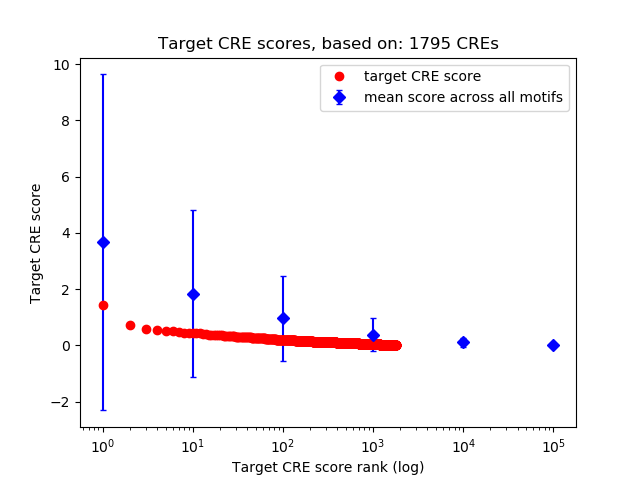
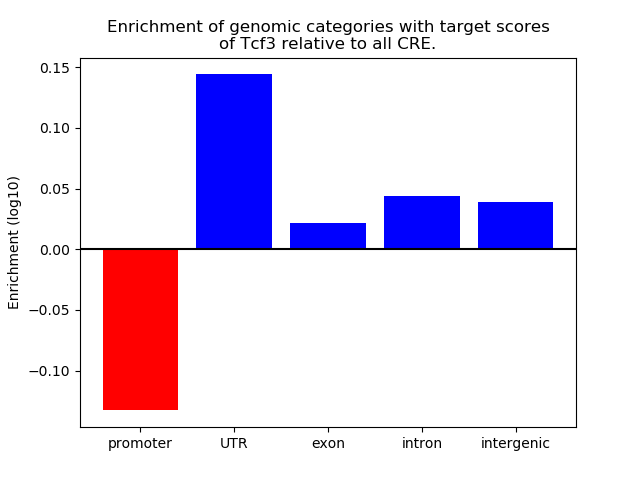
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_19851376_19851638 | 1.43 |
Gm12029 |
predicted gene 12029 |
67855 |
0.1 |
| chr3_89147086_89147561 | 0.73 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
3891 |
0.08 |
| chr12_104087488_104087669 | 0.57 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
6929 |
0.1 |
| chr10_111318102_111318256 | 0.56 |
Gm40761 |
predicted gene, 40761 |
18945 |
0.17 |
| chr10_91261198_91261441 | 0.52 |
Gm18705 |
predicted gene, 18705 |
1975 |
0.31 |
| chr15_3468356_3468528 | 0.51 |
Ghr |
growth hormone receptor |
3202 |
0.36 |
| chr9_74366017_74366190 | 0.47 |
Nr1h2-ps |
nuclear receptor subfamily 1, group H, member 2, pseudogene |
1040 |
0.56 |
| chr19_18146304_18146531 | 0.44 |
Gm18610 |
predicted gene, 18610 |
59378 |
0.14 |
| chr4_10854377_10854556 | 0.43 |
Gm12919 |
predicted gene 12919 |
623 |
0.61 |
| chr15_3513860_3514011 | 0.42 |
Ghr |
growth hormone receptor |
42291 |
0.19 |
| chr16_5140582_5140765 | 0.42 |
Sec14l5 |
SEC14-like lipid binding 5 |
6436 |
0.13 |
| chr9_122850000_122850173 | 0.42 |
Gm47140 |
predicted gene, 47140 |
1668 |
0.22 |
| chr19_20723027_20723214 | 0.41 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
4442 |
0.3 |
| chr8_66494561_66494830 | 0.39 |
Gm32568 |
predicted gene, 32568 |
364 |
0.85 |
| chr11_16816577_16816728 | 0.38 |
Egfros |
epidermal growth factor receptor, opposite strand |
14050 |
0.21 |
| chr11_120819185_120819682 | 0.38 |
Fasn |
fatty acid synthase |
3978 |
0.11 |
| chr7_24919736_24919914 | 0.37 |
Arhgef1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
623 |
0.53 |
| chr2_58769900_58770051 | 0.36 |
Upp2 |
uridine phosphorylase 2 |
4650 |
0.24 |
| chr17_10093572_10093736 | 0.36 |
Qk |
quaking |
119920 |
0.06 |
| chr13_58192282_58192443 | 0.35 |
Gm48357 |
predicted gene, 48357 |
1044 |
0.37 |
| chr2_152359606_152359999 | 0.35 |
Gm14165 |
predicted gene 14165 |
6187 |
0.1 |
| chr11_5904597_5904748 | 0.35 |
Gck |
glucokinase |
2585 |
0.17 |
| chr11_16868479_16868644 | 0.34 |
Egfr |
epidermal growth factor receptor |
9589 |
0.2 |
| chr11_16858660_16858826 | 0.33 |
Egfr |
epidermal growth factor receptor |
19407 |
0.18 |
| chr13_19246519_19246807 | 0.33 |
Trgc3 |
T cell receptor gamma, constant 3 |
14221 |
0.22 |
| chr8_93180612_93180763 | 0.32 |
Ces1d |
carboxylesterase 1D |
5398 |
0.15 |
| chr9_122848440_122848788 | 0.31 |
Gm47140 |
predicted gene, 47140 |
196 |
0.89 |
| chr4_117831574_117831756 | 0.31 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
2841 |
0.16 |
| chr5_130303522_130303878 | 0.31 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
44770 |
0.1 |
| chr11_106412764_106412932 | 0.31 |
Icam2 |
intercellular adhesion molecule 2 |
24773 |
0.12 |
| chr1_50806470_50806812 | 0.31 |
Gm28321 |
predicted gene 28321 |
8734 |
0.28 |
| chr11_22372894_22373087 | 0.31 |
Rpsa-ps3 |
ribosomal protein SA, pseudogene 3 |
13302 |
0.21 |
| chr6_37347891_37348042 | 0.30 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
11431 |
0.26 |
| chr8_120046709_120046884 | 0.30 |
Gm15684 |
predicted gene 15684 |
1509 |
0.32 |
| chr1_59202432_59202591 | 0.29 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
16615 |
0.14 |
| chr11_16833397_16833632 | 0.29 |
Egfros |
epidermal growth factor receptor, opposite strand |
2812 |
0.3 |
| chr10_111314954_111315105 | 0.29 |
Bbs10 |
Bardet-Biedl syndrome 10 (human) |
16350 |
0.18 |
| chr2_49177399_49177550 | 0.28 |
Gm13510 |
predicted gene 13510 |
7544 |
0.27 |
| chr16_49907450_49907601 | 0.28 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
40692 |
0.2 |
| chr19_57506666_57506817 | 0.28 |
6720468P15Rik |
RIKEN cDNA 6720468P15 gene |
1823 |
0.28 |
| chr12_30372401_30372579 | 0.28 |
Sntg2 |
syntrophin, gamma 2 |
775 |
0.73 |
| chr3_87801773_87801942 | 0.28 |
Insrr |
insulin receptor-related receptor |
86 |
0.95 |
| chr15_58960813_58961185 | 0.28 |
Mtss1 |
MTSS I-BAR domain containing 1 |
4159 |
0.17 |
| chr2_101762531_101762726 | 0.28 |
Prr5l |
proline rich 5 like |
35022 |
0.17 |
| chr13_113844856_113845031 | 0.28 |
Arl15 |
ADP-ribosylation factor-like 15 |
50321 |
0.11 |
| chr1_9578333_9578671 | 0.27 |
Gm6161 |
predicted gene 6161 |
3954 |
0.16 |
| chr11_70083308_70083461 | 0.27 |
Asgr2 |
asialoglycoprotein receptor 2 |
9260 |
0.09 |
| chr1_164426310_164426497 | 0.27 |
Gm37411 |
predicted gene, 37411 |
1690 |
0.31 |
| chr15_85233381_85233563 | 0.27 |
Fbln1 |
fibulin 1 |
27467 |
0.17 |
| chr12_108292499_108292690 | 0.27 |
Hhipl1 |
hedgehog interacting protein-like 1 |
13676 |
0.15 |
| chr5_113141351_113141509 | 0.26 |
4930557B06Rik |
RIKEN cDNA 4930557B06 gene |
1019 |
0.36 |
| chr4_102511795_102511975 | 0.26 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
41488 |
0.21 |
| chr6_55108463_55108627 | 0.26 |
Crhr2 |
corticotropin releasing hormone receptor 2 |
7972 |
0.19 |
| chr2_167292312_167292496 | 0.26 |
Gm11473 |
predicted gene 11473 |
10627 |
0.16 |
| chr15_75047474_75047625 | 0.26 |
Ly6c1 |
lymphocyte antigen 6 complex, locus C1 |
1120 |
0.32 |
| chr11_51494380_51494743 | 0.25 |
Col23a1 |
collagen, type XXIII, alpha 1 |
79871 |
0.07 |
| chr5_24502879_24503079 | 0.25 |
Agap3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
22807 |
0.08 |
| chr16_11098511_11098684 | 0.25 |
Txndc11 |
thioredoxin domain containing 11 |
11245 |
0.1 |
| chr10_86872197_86872354 | 0.25 |
Stab2 |
stabilin 2 |
6683 |
0.14 |
| chr2_17326719_17326870 | 0.25 |
Nebl |
nebulette |
34292 |
0.22 |
| chr15_58993520_58993677 | 0.25 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
9462 |
0.17 |
| chr11_22893094_22893344 | 0.25 |
Gm24917 |
predicted gene, 24917 |
24318 |
0.11 |
| chr18_8328204_8328355 | 0.25 |
Gm5500 |
predicted pseudogene 5500 |
86850 |
0.09 |
| chr4_49553429_49553778 | 0.24 |
Aldob |
aldolase B, fructose-bisphosphate |
4057 |
0.17 |
| chr8_46385009_46385281 | 0.24 |
Gm45253 |
predicted gene 45253 |
743 |
0.6 |
| chr10_69208384_69208535 | 0.24 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
93 |
0.97 |
| chr11_110499189_110499340 | 0.24 |
Map2k6 |
mitogen-activated protein kinase kinase 6 |
20225 |
0.24 |
| chr11_98770698_98771034 | 0.23 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
440 |
0.69 |
| chr7_15939282_15939433 | 0.23 |
Snord23 |
small nucleolar RNA, C/D box 23 |
487 |
0.65 |
| chr8_36548724_36548879 | 0.23 |
Dlc1 |
deleted in liver cancer 1 |
28468 |
0.19 |
| chr18_80261215_80262161 | 0.23 |
Slc66a2 |
solute carrier family 66 member 2 |
1152 |
0.35 |
| chr19_46632420_46632695 | 0.23 |
Wbp1l |
WW domain binding protein 1 like |
9156 |
0.13 |
| chr5_130042317_130042468 | 0.23 |
Crcp |
calcitonin gene-related peptide-receptor component protein |
188 |
0.92 |
| chr9_108401502_108401859 | 0.23 |
1700102P08Rik |
RIKEN cDNA 1700102P08 gene |
8846 |
0.07 |
| chr12_85299100_85299299 | 0.23 |
Zc2hc1c |
zinc finger, C2HC-type containing 1C |
10608 |
0.1 |
| chr15_98738867_98739043 | 0.22 |
Fkbp11 |
FK506 binding protein 11 |
10757 |
0.09 |
| chr2_121193827_121193979 | 0.22 |
Tubgcp4 |
tubulin, gamma complex associated protein 4 |
1956 |
0.22 |
| chr17_78241487_78241665 | 0.21 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
3804 |
0.22 |
| chr8_77012726_77012888 | 0.21 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
104535 |
0.06 |
| chr5_88367022_88367405 | 0.21 |
Amtn |
amelotin |
8895 |
0.17 |
| chr4_118002076_118002227 | 0.21 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
27115 |
0.13 |
| chr5_81910056_81910231 | 0.21 |
2900064F13Rik |
RIKEN cDNA 2900064F13 gene |
83327 |
0.09 |
| chr18_64006042_64006212 | 0.21 |
Gm6974 |
predicted gene 6974 |
75293 |
0.09 |
| chr2_20939712_20939881 | 0.21 |
Arhgap21 |
Rho GTPase activating protein 21 |
3623 |
0.24 |
| chr18_67579213_67579374 | 0.21 |
Gm17669 |
predicted gene, 17669 |
16899 |
0.16 |
| chr7_4647276_4647517 | 0.21 |
Ppp6r1 |
protein phosphatase 6, regulatory subunit 1 |
9 |
0.94 |
| chr17_24674148_24674299 | 0.20 |
Zfp598 |
zinc finger protein 598 |
2919 |
0.09 |
| chr11_16818044_16818409 | 0.20 |
Egfros |
epidermal growth factor receptor, opposite strand |
12476 |
0.22 |
| chr18_84376572_84376752 | 0.20 |
Gm37216 |
predicted gene, 37216 |
516 |
0.84 |
| chr11_120248567_120248734 | 0.20 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
11951 |
0.09 |
| chr2_52372456_52372665 | 0.20 |
Neb |
nebulin |
5030 |
0.17 |
| chr11_16798191_16798439 | 0.20 |
Egfros |
epidermal growth factor receptor, opposite strand |
32387 |
0.16 |
| chr10_81378463_81379124 | 0.20 |
Fzr1 |
fizzy and cell division cycle 20 related 1 |
277 |
0.71 |
| chr14_67917259_67917426 | 0.20 |
Dock5 |
dedicator of cytokinesis 5 |
16090 |
0.22 |
| chr13_4270977_4271134 | 0.20 |
Akr1c12 |
aldo-keto reductase family 1, member C12 |
8378 |
0.15 |
| chr13_69631727_69631878 | 0.20 |
Nsun2 |
NOL1/NOP2/Sun domain family member 2 |
1062 |
0.36 |
| chr12_105005460_105005678 | 0.20 |
Syne3 |
spectrin repeat containing, nuclear envelope family member 3 |
4240 |
0.13 |
| chr6_43272880_43273031 | 0.20 |
Arhgef5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
1556 |
0.25 |
| chr9_107649361_107649537 | 0.19 |
Slc38a3 |
solute carrier family 38, member 3 |
6571 |
0.08 |
| chr4_53086731_53086906 | 0.19 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
45892 |
0.13 |
| chr10_4120834_4121002 | 0.19 |
Gm25515 |
predicted gene, 25515 |
16605 |
0.2 |
| chr14_78548923_78549074 | 0.19 |
Akap11 |
A kinase (PRKA) anchor protein 11 |
12190 |
0.2 |
| chr7_75611829_75612100 | 0.19 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
124 |
0.97 |
| chr15_59020453_59020630 | 0.19 |
Mtss1 |
MTSS I-BAR domain containing 1 |
20055 |
0.17 |
| chr13_96660106_96660425 | 0.19 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
3598 |
0.18 |
| chr4_49538717_49538957 | 0.19 |
Aldob |
aldolase B, fructose-bisphosphate |
29 |
0.97 |
| chr2_172938722_172938940 | 0.19 |
Bmp7 |
bone morphogenetic protein 7 |
1261 |
0.45 |
| chr3_75868999_75869188 | 0.19 |
Golim4 |
golgi integral membrane protein 4 |
9915 |
0.24 |
| chr18_76254679_76254860 | 0.19 |
Smad2 |
SMAD family member 2 |
6352 |
0.2 |
| chr6_38823949_38824290 | 0.19 |
Hipk2 |
homeodomain interacting protein kinase 2 |
5773 |
0.25 |
| chr19_37433970_37434137 | 0.19 |
Gm38345 |
predicted gene, 38345 |
603 |
0.45 |
| chr17_80697982_80698359 | 0.19 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
29854 |
0.18 |
| chr5_23462026_23462363 | 0.19 |
Kmt2e |
lysine (K)-specific methyltransferase 2E |
16361 |
0.15 |
| chr15_9079456_9079615 | 0.18 |
Nadk2 |
NAD kinase 2, mitochondrial |
4258 |
0.25 |
| chr16_10366678_10366849 | 0.18 |
Gm1600 |
predicted gene 1600 |
19172 |
0.14 |
| chr14_24960151_24960328 | 0.18 |
Gm10398 |
predicted gene 10398 |
50825 |
0.13 |
| chr11_16769921_16770266 | 0.18 |
Egfr |
epidermal growth factor receptor |
17863 |
0.19 |
| chr7_31101720_31101871 | 0.18 |
Hpn |
hepsin |
719 |
0.46 |
| chr2_32409058_32409478 | 0.18 |
Gm13413 |
predicted gene 13413 |
3213 |
0.12 |
| chr12_103670437_103670601 | 0.18 |
Serpina16 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
4945 |
0.12 |
| chr1_126484684_126484835 | 0.18 |
Nckap5 |
NCK-associated protein 5 |
7983 |
0.32 |
| chr2_127367630_127367825 | 0.18 |
Adra2b |
adrenergic receptor, alpha 2b |
4441 |
0.16 |
| chr4_130708524_130708794 | 0.18 |
Snord85 |
small nucleolar RNA, C/D box 85 |
40975 |
0.1 |
| chr9_57214145_57214320 | 0.18 |
Trcg1 |
taste receptor cell gene 1 |
22324 |
0.12 |
| chr16_30664084_30664254 | 0.18 |
Fam43a |
family with sequence similarity 43, member A |
64446 |
0.1 |
| chr1_62809972_62810173 | 0.18 |
Gm37121 |
predicted gene, 37121 |
770 |
0.67 |
| chr12_32707351_32707528 | 0.18 |
Gm18726 |
predicted gene, 18726 |
2722 |
0.31 |
| chr2_134826467_134826626 | 0.17 |
Gm14036 |
predicted gene 14036 |
22597 |
0.2 |
| chr3_24630236_24630408 | 0.17 |
Gm24704 |
predicted gene, 24704 |
68248 |
0.14 |
| chr6_84235021_84235302 | 0.17 |
Gm44127 |
predicted gene, 44127 |
9909 |
0.22 |
| chr15_99101408_99101603 | 0.17 |
Dnajc22 |
DnaJ heat shock protein family (Hsp40) member C22 |
2093 |
0.16 |
| chr8_111739071_111739223 | 0.17 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
4662 |
0.21 |
| chr4_86373682_86373833 | 0.17 |
Gm12550 |
predicted gene 12550 |
68584 |
0.11 |
| chr15_10206547_10206755 | 0.17 |
Prlr |
prolactin receptor |
6814 |
0.3 |
| chr2_174330237_174330577 | 0.17 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
120 |
0.95 |
| chr2_124609977_124610547 | 0.17 |
Sema6d |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
34 |
0.99 |
| chr8_26078485_26078636 | 0.17 |
Hook3 |
hook microtubule tethering protein 3 |
817 |
0.46 |
| chr15_76227819_76227970 | 0.17 |
Plec |
plectin |
1603 |
0.17 |
| chr13_108042230_108042425 | 0.17 |
Smim15 |
small integral membrane protein 15 |
2119 |
0.37 |
| chr9_67137735_67137898 | 0.17 |
Gm19299 |
predicted gene, 19299 |
4255 |
0.21 |
| chr11_59226278_59226466 | 0.17 |
Arf1 |
ADP-ribosylation factor 1 |
1790 |
0.2 |
| chr16_26669940_26670091 | 0.17 |
Il1rap |
interleukin 1 receptor accessory protein |
45859 |
0.18 |
| chr7_68728361_68728534 | 0.16 |
Gm44692 |
predicted gene 44692 |
1980 |
0.37 |
| chr5_135703012_135703168 | 0.16 |
Gm16061 |
predicted gene 16061 |
4071 |
0.13 |
| chr5_36176782_36177189 | 0.16 |
Psapl1 |
prosaposin-like 1 |
27036 |
0.21 |
| chr4_139454119_139454298 | 0.16 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
9051 |
0.18 |
| chr6_38870157_38870322 | 0.16 |
Tbxas1 |
thromboxane A synthase 1, platelet |
5165 |
0.22 |
| chr19_9027860_9028087 | 0.16 |
Ahnak |
AHNAK nucleoprotein (desmoyokin) |
28313 |
0.09 |
| chr17_35865754_35865905 | 0.16 |
Ppp1r18 |
protein phosphatase 1, regulatory subunit 18 |
204 |
0.76 |
| chr8_11145382_11145560 | 0.16 |
Gm44717 |
predicted gene 44717 |
3775 |
0.2 |
| chr14_25508473_25508633 | 0.16 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
5898 |
0.16 |
| chr1_130387327_130387514 | 0.16 |
Cd55b |
CD55 molecule, decay accelerating factor for complement B |
35320 |
0.11 |
| chr15_59014305_59014721 | 0.16 |
Mtss1 |
MTSS I-BAR domain containing 1 |
26083 |
0.16 |
| chr3_107230665_107230816 | 0.16 |
A630076J17Rik |
RIKEN cDNA A630076J17 gene |
126 |
0.94 |
| chr5_51144711_51144875 | 0.16 |
Gm44377 |
predicted gene, 44377 |
78881 |
0.1 |
| chr7_79308613_79308815 | 0.16 |
Gm39041 |
predicted gene, 39041 |
9689 |
0.14 |
| chr6_51573063_51573238 | 0.16 |
Snx10 |
sorting nexin 10 |
28584 |
0.17 |
| chr15_89102253_89102404 | 0.16 |
Tubgcp6 |
tubulin, gamma complex associated protein 6 |
44 |
0.94 |
| chr15_55843743_55843894 | 0.16 |
Sntb1 |
syntrophin, basic 1 |
62492 |
0.11 |
| chr6_136906748_136907166 | 0.16 |
Erp27 |
endoplasmic reticulum protein 27 |
15183 |
0.11 |
| chr14_67884776_67884962 | 0.16 |
Mir6539 |
microRNA 6539 |
25124 |
0.2 |
| chr3_129557154_129557334 | 0.16 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
4864 |
0.19 |
| chr14_16478758_16478933 | 0.16 |
Top2b |
topoisomerase (DNA) II beta |
53417 |
0.13 |
| chr15_83772573_83772737 | 0.15 |
Mpped1 |
metallophosphoesterase domain containing 1 |
6812 |
0.24 |
| chr17_47593664_47594464 | 0.15 |
Ccnd3 |
cyclin D3 |
249 |
0.86 |
| chr12_110221880_110222031 | 0.15 |
Gm40576 |
predicted gene, 40576 |
14407 |
0.11 |
| chr11_117233984_117234135 | 0.15 |
Septin9 |
septin 9 |
1774 |
0.34 |
| chr11_50104096_50104296 | 0.15 |
Rnf130 |
ring finger protein 130 |
10522 |
0.12 |
| chr11_6016490_6016658 | 0.15 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
16126 |
0.16 |
| chr5_138948306_138948628 | 0.15 |
Pdgfa |
platelet derived growth factor, alpha |
45814 |
0.12 |
| chr19_48703871_48704022 | 0.15 |
Sorcs3 |
sortilin-related VPS10 domain containing receptor 3 |
270 |
0.95 |
| chr16_92637112_92637263 | 0.15 |
Gm26626 |
predicted gene, 26626 |
24373 |
0.19 |
| chr13_9199577_9199754 | 0.15 |
Gm8784 |
predicted gene 8784 |
23872 |
0.13 |
| chr8_71495277_71495471 | 0.15 |
Gtpbp3 |
GTP binding protein 3 |
5915 |
0.08 |
| chr5_134209731_134209889 | 0.15 |
Gtf2ird2 |
GTF2I repeat domain containing 2 |
3153 |
0.17 |
| chr14_60976604_60976847 | 0.15 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
840 |
0.7 |
| chr10_68103006_68103157 | 0.15 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
33545 |
0.18 |
| chr3_138280557_138280953 | 0.15 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
3104 |
0.16 |
| chr10_11306105_11306256 | 0.15 |
Gm48666 |
predicted gene, 48666 |
24235 |
0.12 |
| chr16_95851988_95852139 | 0.15 |
1600002D24Rik |
RIKEN cDNA 1600002D24 gene |
6293 |
0.2 |
| chr10_117884880_117885031 | 0.15 |
Gm32552 |
predicted gene, 32552 |
18240 |
0.14 |
| chr7_137286258_137286429 | 0.15 |
Ebf3 |
early B cell factor 3 |
18805 |
0.18 |
| chr1_73513962_73514123 | 0.15 |
Gm9553 |
predicted gene 9553 |
1996 |
0.31 |
| chr1_67197545_67197701 | 0.15 |
Gm15668 |
predicted gene 15668 |
51577 |
0.13 |
| chr6_67036040_67036201 | 0.15 |
E230016M11Rik |
RIKEN cDNA E230016M11 gene |
479 |
0.61 |
| chr7_99165012_99165417 | 0.15 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
4892 |
0.14 |
| chr2_18159543_18159694 | 0.14 |
Gm25147 |
predicted gene, 25147 |
931 |
0.48 |
| chr11_22494745_22494936 | 0.14 |
Gm12051 |
predicted gene 12051 |
11232 |
0.19 |
| chr19_53786299_53786461 | 0.14 |
Rbm20 |
RNA binding motif protein 20 |
6928 |
0.18 |
| chr6_139624876_139625027 | 0.14 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
3063 |
0.3 |
| chr15_64362676_64362949 | 0.14 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
12926 |
0.19 |
| chr19_29862924_29863094 | 0.14 |
Gm50380 |
predicted gene, 50380 |
12208 |
0.18 |
| chr6_101046905_101047082 | 0.14 |
Gm43948 |
predicted gene, 43948 |
12178 |
0.18 |
| chr7_119523427_119523676 | 0.14 |
Pdilt |
protein disulfide isomerase-like, testis expressed |
62 |
0.95 |
| chr3_41498956_41499107 | 0.14 |
Platr4 |
pluripotency associated transcript 4 |
5839 |
0.14 |
| chr10_69257688_69257866 | 0.14 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
7860 |
0.2 |
| chr12_103805644_103805795 | 0.14 |
Gm17043 |
predicted gene 17043 |
1357 |
0.28 |
| chr2_27767443_27767901 | 0.14 |
Rxra |
retinoid X receptor alpha |
27471 |
0.21 |
| chr1_181529523_181529674 | 0.14 |
Ccdc121 |
coiled-coil domain containing 121 |
18147 |
0.19 |
| chr1_157101218_157101391 | 0.14 |
Tex35 |
testis expressed 35 |
7084 |
0.18 |
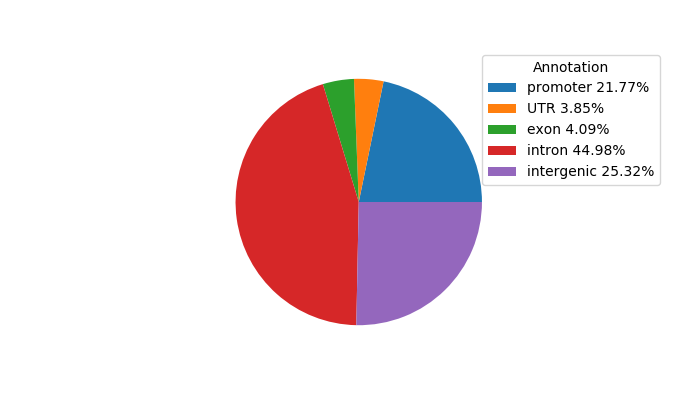
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0044805 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.0 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.0 | GO:0035793 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |