Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
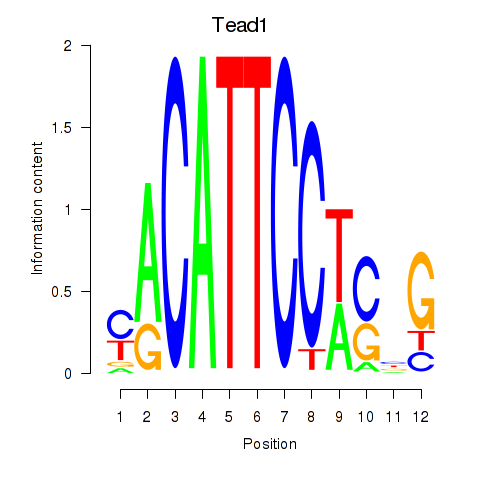
Results for Tead1
Z-value: 1.87
Transcription factors associated with Tead1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead1
|
ENSMUSG00000055320.10 | TEA domain family member 1 |
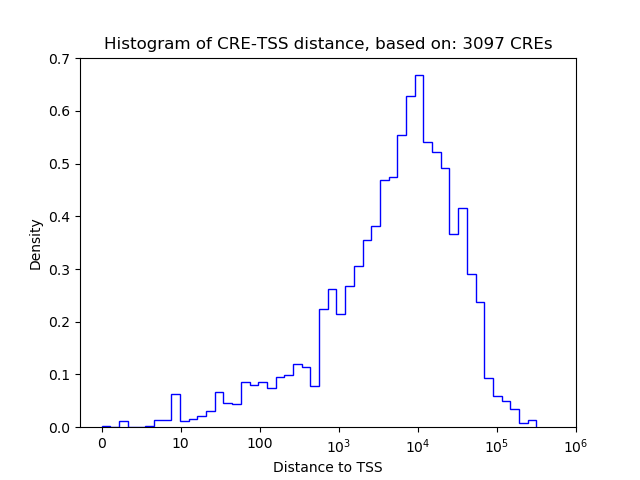
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_112697188_112697354 | Tead1 | 5573 | 0.180704 | 0.88 | 1.9e-02 | Click! |
| chr7_112697541_112697813 | Tead1 | 5979 | 0.178198 | 0.87 | 2.3e-02 | Click! |
| chr7_112688480_112688662 | Tead1 | 3127 | 0.214614 | 0.86 | 2.6e-02 | Click! |
| chr7_112807573_112807746 | Tead1 | 31975 | 0.191092 | 0.78 | 6.9e-02 | Click! |
| chr7_112788724_112788885 | Tead1 | 29258 | 0.192438 | 0.77 | 7.5e-02 | Click! |
Activity of the Tead1 motif across conditions
Conditions sorted by the z-value of the Tead1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
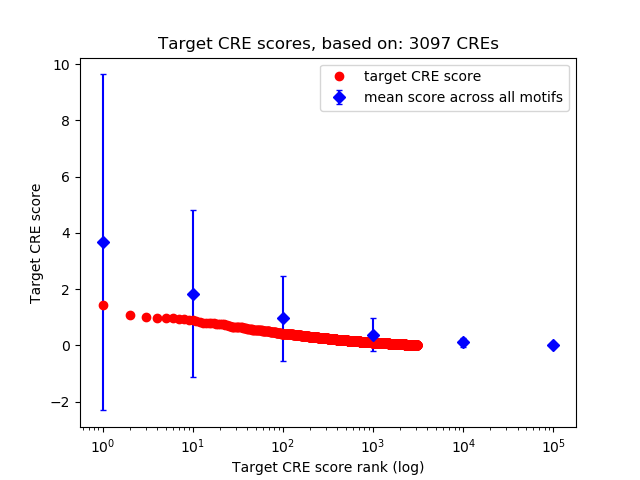
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_24595287_24595581 | 1.44 |
Ccn2 |
cellular communication network factor 2 |
8 |
0.86 |
| chr7_84457901_84458052 | 1.07 |
Gm45175 |
predicted gene 45175 |
13425 |
0.16 |
| chr5_139317463_139317630 | 1.00 |
Adap1 |
ArfGAP with dual PH domains 1 |
8076 |
0.13 |
| chr11_88173046_88173197 | 0.98 |
Cuedc1 |
CUE domain containing 1 |
9182 |
0.18 |
| chr5_151412960_151413135 | 0.98 |
1700028E10Rik |
RIKEN cDNA 1700028E10 gene |
6521 |
0.17 |
| chr12_8333733_8333892 | 0.96 |
Gm48071 |
predicted gene, 48071 |
2905 |
0.21 |
| chr10_127544019_127544184 | 0.95 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
893 |
0.41 |
| chr5_144258484_144258651 | 0.94 |
2900089D17Rik |
RIKEN cDNA 2900089D17 gene |
2381 |
0.19 |
| chr3_88102838_88102989 | 0.89 |
Iqgap3 |
IQ motif containing GTPase activating protein 3 |
2229 |
0.18 |
| chr17_35549631_35549795 | 0.89 |
Cdsn |
corneodesmosin |
2415 |
0.12 |
| chr7_45710445_45711101 | 0.86 |
Sphk2 |
sphingosine kinase 2 |
2670 |
0.09 |
| chr15_99041222_99041434 | 0.83 |
Tuba1c |
tubulin, alpha 1C |
11007 |
0.09 |
| chr15_62501057_62501225 | 0.81 |
Gm41333 |
predicted gene, 41333 |
66834 |
0.14 |
| chr2_25176703_25176854 | 0.81 |
Nrarp |
Notch-regulated ankyrin repeat protein |
3980 |
0.08 |
| chr7_110297212_110297456 | 0.80 |
Gm9132 |
predicted gene 9132 |
4667 |
0.2 |
| chr7_45705992_45706174 | 0.79 |
Dbp |
D site albumin promoter binding protein |
799 |
0.31 |
| chr4_117997646_117997813 | 0.78 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
22693 |
0.14 |
| chr3_152143501_152143652 | 0.77 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
3212 |
0.17 |
| chr13_69524173_69524569 | 0.77 |
Tent4a |
terminal nucleotidyltransferase 4A |
8061 |
0.13 |
| chr14_7874610_7874781 | 0.77 |
Flnb |
filamin, beta |
10046 |
0.2 |
| chr4_128946455_128946665 | 0.74 |
Gm15904 |
predicted gene 15904 |
10555 |
0.15 |
| chr15_98868738_98868889 | 0.74 |
Kmt2d |
lysine (K)-specific methyltransferase 2D |
2370 |
0.12 |
| chr15_93916844_93917358 | 0.73 |
9430014N10Rik |
RIKEN cDNA 9430014N10 gene |
10010 |
0.25 |
| chr17_5075206_5075364 | 0.71 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
34707 |
0.18 |
| chr13_112405475_112405627 | 0.69 |
2810403G07Rik |
RIKEN cDNA 2810403G07 gene |
11814 |
0.14 |
| chr10_20749070_20749231 | 0.67 |
Gm48651 |
predicted gene, 48651 |
6967 |
0.23 |
| chr2_29731675_29731853 | 0.67 |
Rapgef1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
797 |
0.51 |
| chr1_182670647_182670811 | 0.66 |
Gm37516 |
predicted gene, 37516 |
9210 |
0.19 |
| chr15_97773204_97773381 | 0.66 |
Slc48a1 |
solute carrier family 48 (heme transporter), member 1 |
5228 |
0.13 |
| chr14_27350759_27350910 | 0.66 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
10555 |
0.2 |
| chr1_40217644_40217810 | 0.66 |
Il1r1 |
interleukin 1 receptor, type I |
7353 |
0.21 |
| chr18_64381354_64381662 | 0.65 |
Onecut2 |
one cut domain, family member 2 |
41488 |
0.11 |
| chr5_148998636_148998818 | 0.64 |
Gm15406 |
predicted gene 15406 |
2161 |
0.14 |
| chr17_47193022_47193193 | 0.64 |
Trerf1 |
transcriptional regulating factor 1 |
19280 |
0.21 |
| chr11_108198624_108198935 | 0.63 |
Gm11655 |
predicted gene 11655 |
16929 |
0.25 |
| chr4_135820255_135820445 | 0.63 |
Myom3 |
myomesin family, member 3 |
19706 |
0.11 |
| chr5_139395256_139395425 | 0.63 |
Gpr146 |
G protein-coupled receptor 146 |
5555 |
0.12 |
| chr18_61360632_61360830 | 0.61 |
Gm25301 |
predicted gene, 25301 |
37082 |
0.11 |
| chr14_65863199_65863389 | 0.60 |
Ccdc25 |
coiled-coil domain containing 25 |
25934 |
0.15 |
| chr11_60249007_60249158 | 0.60 |
Tom1l2 |
target of myb1-like 2 (chicken) |
3413 |
0.16 |
| chr1_13103635_13103786 | 0.58 |
Prdm14 |
PR domain containing 14 |
23453 |
0.13 |
| chr9_15525292_15525443 | 0.58 |
Smco4 |
single-pass membrane protein with coiled-coil domains 4 |
4508 |
0.2 |
| chr7_34319277_34319447 | 0.57 |
4931406P16Rik |
RIKEN cDNA 4931406P16 gene |
5811 |
0.13 |
| chr19_10054766_10055233 | 0.57 |
Fads3 |
fatty acid desaturase 3 |
1253 |
0.35 |
| chr4_105179379_105179579 | 0.56 |
Plpp3 |
phospholipid phosphatase 3 |
22132 |
0.24 |
| chr2_25145394_25145761 | 0.55 |
Gm13387 |
predicted gene 13387 |
181 |
0.87 |
| chrX_169054094_169054245 | 0.55 |
Arhgap6 |
Rho GTPase activating protein 6 |
17558 |
0.24 |
| chr3_121982496_121982688 | 0.55 |
Arhgap29 |
Rho GTPase activating protein 29 |
1042 |
0.54 |
| chr17_78206502_78206668 | 0.55 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
5520 |
0.16 |
| chr2_178993954_178994105 | 0.55 |
Gm14314 |
predicted gene 14314 |
27104 |
0.26 |
| chr17_84074449_84074742 | 0.54 |
4933433H22Rik |
RIKEN cDNA 4933433H22 gene |
4065 |
0.18 |
| chr6_28681979_28682177 | 0.54 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
22831 |
0.21 |
| chr1_57890324_57890494 | 0.54 |
Spats2l |
spermatogenesis associated, serine-rich 2-like |
11662 |
0.2 |
| chr3_84298294_84298452 | 0.53 |
Trim2 |
tripartite motif-containing 2 |
6368 |
0.27 |
| chr14_54572931_54573082 | 0.53 |
Ajuba |
ajuba LIM protein |
924 |
0.35 |
| chr1_184072575_184072786 | 0.53 |
Dusp10 |
dual specificity phosphatase 10 |
38299 |
0.17 |
| chr18_35830960_35831336 | 0.53 |
Gm29417 |
predicted gene 29417 |
55 |
0.92 |
| chr12_37050724_37050882 | 0.53 |
Gm48617 |
predicted gene, 48617 |
1586 |
0.37 |
| chr11_16588986_16589143 | 0.52 |
Gm12663 |
predicted gene 12663 |
47002 |
0.12 |
| chr11_87735822_87735981 | 0.52 |
Supt4a |
SPT4A, DSIF elongation factor subunit |
1651 |
0.19 |
| chr7_89558667_89558818 | 0.51 |
Gm45064 |
predicted gene 45064 |
6376 |
0.17 |
| chr13_44409176_44409378 | 0.51 |
Gm33958 |
predicted gene, 33958 |
18160 |
0.15 |
| chr10_22875325_22875476 | 0.51 |
Tcf21 |
transcription factor 21 |
55226 |
0.11 |
| chr1_184069803_184070002 | 0.51 |
Dusp10 |
dual specificity phosphatase 10 |
35521 |
0.18 |
| chr7_132723077_132723229 | 0.51 |
Fam53b |
family with sequence similarity 53, member B |
53763 |
0.11 |
| chr7_34485188_34485527 | 0.50 |
Gm12780 |
predicted gene 12780 |
10708 |
0.16 |
| chr15_83440834_83440989 | 0.50 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
8087 |
0.16 |
| chr12_111691358_111691542 | 0.50 |
Gm36635 |
predicted gene, 36635 |
11632 |
0.09 |
| chr10_20669582_20669754 | 0.49 |
Gm17230 |
predicted gene 17230 |
44033 |
0.15 |
| chr8_128447394_128447576 | 0.49 |
Nrp1 |
neuropilin 1 |
88088 |
0.08 |
| chr6_35875002_35875227 | 0.49 |
Gm43442 |
predicted gene 43442 |
51597 |
0.17 |
| chr17_31965712_31965881 | 0.49 |
Gm17572 |
predicted gene, 17572 |
15262 |
0.15 |
| chr10_17786858_17787009 | 0.48 |
Gm20655 |
predicted gene 20655 |
8395 |
0.18 |
| chr15_75270297_75270448 | 0.48 |
Ly6f |
lymphocyte antigen 6 complex, locus F |
1670 |
0.23 |
| chr13_115090695_115090877 | 0.48 |
Pelo |
pelota mRNA surveillance and ribosome rescue factor |
600 |
0.58 |
| chr2_80038295_80038479 | 0.48 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
642 |
0.82 |
| chr9_108600199_108600350 | 0.47 |
P4htm |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
2607 |
0.09 |
| chr5_121686462_121686752 | 0.47 |
Brap |
BRCA1 associated protein |
3679 |
0.14 |
| chr15_75593070_75593251 | 0.46 |
Gpihbp1 |
GPI-anchored HDL-binding protein 1 |
3468 |
0.15 |
| chr11_75134022_75134199 | 0.46 |
Smg6 |
Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) |
1675 |
0.24 |
| chr15_96674208_96674363 | 0.46 |
Gm22045 |
predicted gene, 22045 |
2492 |
0.26 |
| chr12_86948719_86948887 | 0.46 |
Cipc |
CLOCK interacting protein, circadian |
1313 |
0.41 |
| chr19_29143356_29143666 | 0.46 |
Mir101b |
microRNA 101b |
8232 |
0.15 |
| chr6_137142914_137143065 | 0.45 |
Rerg |
RAS-like, estrogen-regulated, growth-inhibitor |
2926 |
0.27 |
| chr17_79451613_79451775 | 0.45 |
Cdc42ep3 |
CDC42 effector protein (Rho GTPase binding) 3 |
96603 |
0.07 |
| chr7_65469723_65469896 | 0.45 |
Gm44792 |
predicted gene 44792 |
11412 |
0.21 |
| chr11_82033062_82033234 | 0.45 |
Ccl2 |
chemokine (C-C motif) ligand 2 |
2423 |
0.18 |
| chr2_58765113_58765779 | 0.45 |
Upp2 |
uridine phosphorylase 2 |
121 |
0.97 |
| chr16_16224499_16224674 | 0.44 |
Pkp2 |
plakophilin 2 |
11268 |
0.19 |
| chrX_129413614_129413812 | 0.43 |
Gm14986 |
predicted gene 14986 |
104813 |
0.07 |
| chr12_58323949_58324220 | 0.43 |
Clec14a |
C-type lectin domain family 14, member a |
54794 |
0.16 |
| chr8_70665733_70665894 | 0.43 |
Pgpep1 |
pyroglutamyl-peptidase I |
5425 |
0.09 |
| chr9_31223628_31223791 | 0.43 |
Aplp2 |
amyloid beta (A4) precursor-like protein 2 |
11894 |
0.18 |
| chr6_28704741_28704911 | 0.42 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
83 |
0.98 |
| chr11_102221971_102222363 | 0.42 |
Hdac5 |
histone deacetylase 5 |
3238 |
0.12 |
| chr5_53193460_53193736 | 0.42 |
Sel1l3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
19717 |
0.18 |
| chr2_60698236_60698402 | 0.42 |
Itgb6 |
integrin beta 6 |
7259 |
0.25 |
| chr3_82054654_82054805 | 0.42 |
Gucy1b1 |
guanylate cyclase 1, soluble, beta 1 |
19407 |
0.17 |
| chr19_40241408_40241559 | 0.41 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
10678 |
0.15 |
| chr8_126584050_126584216 | 0.41 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
9853 |
0.22 |
| chr7_57560848_57561014 | 0.41 |
Mir6389 |
microRNA 6389 |
20130 |
0.19 |
| chr1_184852414_184852593 | 0.41 |
Mtarc2 |
mitochondrial amidoxime reducing component 2 |
6052 |
0.17 |
| chr15_64362676_64362949 | 0.41 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
12926 |
0.19 |
| chr9_90260660_90260839 | 0.41 |
Tbc1d2b |
TBC1 domain family, member 2B |
4822 |
0.19 |
| chr7_31108093_31108358 | 0.41 |
Hpn |
hepsin |
2772 |
0.13 |
| chr11_8484705_8484960 | 0.41 |
Tns3 |
tensin 3 |
16157 |
0.28 |
| chr3_10211563_10211714 | 0.40 |
Fabp4 |
fatty acid binding protein 4, adipocyte |
3062 |
0.13 |
| chr7_132736194_132736348 | 0.40 |
Fam53b |
family with sequence similarity 53, member B |
40645 |
0.13 |
| chr7_99168662_99168821 | 0.40 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
1365 |
0.33 |
| chr9_46049413_46049714 | 0.40 |
Sik3 |
SIK family kinase 3 |
36563 |
0.11 |
| chr1_191075136_191075311 | 0.40 |
Gm38188 |
predicted gene, 38188 |
3457 |
0.13 |
| chr2_127603534_127603695 | 0.40 |
Mrps5 |
mitochondrial ribosomal protein S5 |
3581 |
0.16 |
| chr10_77448892_77449069 | 0.40 |
Gm35920 |
predicted gene, 35920 |
8937 |
0.17 |
| chr16_52267318_52267489 | 0.40 |
Alcam |
activated leukocyte cell adhesion molecule |
1658 |
0.53 |
| chr17_75465392_75465653 | 0.40 |
Rasgrp3 |
RAS, guanyl releasing protein 3 |
36 |
0.99 |
| chr10_117729488_117729656 | 0.40 |
Slc35e3 |
solute carrier family 35, member E3 |
15370 |
0.13 |
| chr9_35039370_35039714 | 0.39 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
16252 |
0.16 |
| chr4_149923922_149924117 | 0.39 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
1548 |
0.34 |
| chr3_151880053_151880221 | 0.39 |
Gm43617 |
predicted gene 43617 |
7166 |
0.17 |
| chr12_112587593_112587810 | 0.39 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
1083 |
0.43 |
| chr5_138979896_138980396 | 0.39 |
Pdgfa |
platelet derived growth factor, alpha |
14135 |
0.17 |
| chr17_57224886_57225407 | 0.39 |
C3 |
complement component 3 |
2319 |
0.17 |
| chr6_113668529_113668873 | 0.39 |
Irak2 |
interleukin-1 receptor-associated kinase 2 |
149 |
0.89 |
| chr17_4479666_4479822 | 0.39 |
4930517M08Rik |
RIKEN cDNA 4930517M08 gene |
155234 |
0.04 |
| chr6_89345028_89345199 | 0.39 |
Gm44207 |
predicted gene, 44207 |
27 |
0.97 |
| chr5_142440070_142440526 | 0.39 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
23646 |
0.17 |
| chr10_37250196_37250360 | 0.39 |
4930543K20Rik |
RIKEN cDNA 4930543K20 gene |
13470 |
0.25 |
| chr4_95389795_95389977 | 0.39 |
Gm29064 |
predicted gene 29064 |
12904 |
0.24 |
| chr11_121520060_121520211 | 0.39 |
Zfp750 |
zinc finger protein 750 |
802 |
0.62 |
| chr5_24969338_24969503 | 0.38 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
16422 |
0.17 |
| chr4_106590222_106590393 | 0.38 |
Gm12744 |
predicted gene 12744 |
1199 |
0.34 |
| chr5_22431872_22432035 | 0.38 |
n-R5s170 |
nuclear encoded rRNA 5S 170 |
1510 |
0.34 |
| chr9_116873050_116873366 | 0.38 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
50389 |
0.18 |
| chr2_124753827_124753978 | 0.38 |
Gm13994 |
predicted gene 13994 |
45247 |
0.19 |
| chr7_35146051_35146625 | 0.38 |
Gm35665 |
predicted gene, 35665 |
4889 |
0.12 |
| chr9_63959217_63959382 | 0.38 |
Gm36033 |
predicted gene, 36033 |
22178 |
0.16 |
| chr15_84193628_84193804 | 0.38 |
Samm50 |
SAMM50 sorting and assembly machinery component |
1475 |
0.26 |
| chr8_91377438_91377598 | 0.38 |
Fto |
fat mass and obesity associated |
13037 |
0.15 |
| chr11_102430263_102430421 | 0.38 |
Grn |
granulin |
20 |
0.95 |
| chr8_122461364_122461539 | 0.37 |
Snai3 |
snail family zinc finger 3 |
718 |
0.43 |
| chr3_129551702_129551883 | 0.37 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
588 |
0.72 |
| chr15_64345201_64345377 | 0.37 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
4597 |
0.21 |
| chr1_61971987_61972299 | 0.37 |
Gm29640 |
predicted gene 29640 |
9064 |
0.27 |
| chr3_82952346_82952497 | 0.37 |
Gm30097 |
predicted gene, 30097 |
43164 |
0.1 |
| chr9_4380683_4380845 | 0.36 |
Msantd4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
2771 |
0.26 |
| chr4_150707281_150707454 | 0.36 |
Gm16079 |
predicted gene 16079 |
28575 |
0.17 |
| chr16_50753691_50753842 | 0.36 |
Dubr |
Dppa2 upstream binding RNA |
20993 |
0.17 |
| chr10_28395793_28396016 | 0.36 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
164247 |
0.04 |
| chr11_6021859_6022054 | 0.35 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
21508 |
0.15 |
| chr5_89422485_89422688 | 0.35 |
Gc |
vitamin D binding protein |
13042 |
0.26 |
| chr4_139580884_139581199 | 0.35 |
Iffo2 |
intermediate filament family orphan 2 |
6321 |
0.15 |
| chr4_70521535_70521760 | 0.35 |
Megf9 |
multiple EGF-like-domains 9 |
13281 |
0.3 |
| chr10_86161840_86162013 | 0.35 |
Gm15990 |
predicted gene 15990 |
36099 |
0.14 |
| chr14_116448226_116448591 | 0.35 |
Gm38045 |
predicted gene, 38045 |
301807 |
0.01 |
| chr16_34937792_34937966 | 0.35 |
1700119H24Rik |
RIKEN cDNA 1700119H24 gene |
1450 |
0.39 |
| chr8_35920405_35920556 | 0.35 |
5430403N17Rik |
RIKEN cDNA 5430403N17 gene |
37523 |
0.14 |
| chr6_134504999_134505166 | 0.35 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
33789 |
0.15 |
| chr9_71974657_71974812 | 0.35 |
Gm37663 |
predicted gene, 37663 |
16537 |
0.1 |
| chr9_74873778_74874078 | 0.35 |
Onecut1 |
one cut domain, family member 1 |
7444 |
0.16 |
| chr10_77379991_77380173 | 0.35 |
Adarb1 |
adenosine deaminase, RNA-specific, B1 |
9841 |
0.2 |
| chr1_82258433_82258589 | 0.35 |
Gm9747 |
predicted gene 9747 |
25399 |
0.16 |
| chr1_155202064_155202230 | 0.35 |
Stx6 |
syntaxin 6 |
7126 |
0.15 |
| chr13_101718024_101718189 | 0.34 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
16002 |
0.21 |
| chr12_70050792_70050966 | 0.34 |
Nin |
ninein |
405 |
0.82 |
| chr1_167755293_167755461 | 0.34 |
Lmx1a |
LIM homeobox transcription factor 1 alpha |
65820 |
0.13 |
| chr16_10976433_10976649 | 0.34 |
Litaf |
LPS-induced TN factor |
962 |
0.41 |
| chr18_65222814_65223030 | 0.34 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
15038 |
0.15 |
| chr13_38150631_38150801 | 0.34 |
Dsp |
desmoplakin |
578 |
0.6 |
| chr9_121415708_121416155 | 0.33 |
Trak1 |
trafficking protein, kinesin binding 1 |
44 |
0.98 |
| chr3_107850765_107850942 | 0.33 |
Gm36211 |
predicted gene, 36211 |
17314 |
0.08 |
| chr7_14428724_14428875 | 0.33 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
7648 |
0.17 |
| chr7_72998547_72998698 | 0.33 |
Gm5335 |
predicted gene 5335 |
8469 |
0.25 |
| chr2_122256897_122257048 | 0.33 |
Sord |
sorbitol dehydrogenase |
22223 |
0.09 |
| chr2_126916345_126916504 | 0.33 |
Sppl2a |
signal peptide peptidase like 2A |
3921 |
0.19 |
| chr8_13490011_13490371 | 0.33 |
Gas6 |
growth arrest specific 6 |
4299 |
0.18 |
| chr6_136461910_136462080 | 0.33 |
Gm6728 |
predicted gene 6728 |
25191 |
0.11 |
| chr1_41543586_41543980 | 0.33 |
Gm28634 |
predicted gene 28634 |
14240 |
0.31 |
| chr2_136911982_136912324 | 0.32 |
Slx4ip |
SLX4 interacting protein |
6610 |
0.18 |
| chr16_57582101_57582276 | 0.32 |
Cmss1 |
cms small ribosomal subunit 1 |
24588 |
0.2 |
| chr5_88797033_88797476 | 0.32 |
Gm42912 |
predicted gene 42912 |
3358 |
0.19 |
| chr5_17854594_17854745 | 0.32 |
Cd36 |
CD36 molecule |
4877 |
0.33 |
| chr3_69139928_69140109 | 0.32 |
Gm1647 |
predicted gene 1647 |
8785 |
0.14 |
| chr4_41527930_41528260 | 0.32 |
Fam219a |
family with sequence similarity 219, member A |
3325 |
0.13 |
| chr8_46804538_46804714 | 0.32 |
Irf2 |
interferon regulatory factor 2 |
1698 |
0.32 |
| chr10_20548960_20549132 | 0.32 |
Pde7b |
phosphodiesterase 7B |
576 |
0.79 |
| chrX_60288366_60288543 | 0.32 |
Atp11c |
ATPase, class VI, type 11C |
21624 |
0.19 |
| chr17_24418727_24418886 | 0.32 |
Rnps1 |
RNA binding protein with serine rich domain 1 |
314 |
0.75 |
| chr8_35654448_35654627 | 0.32 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
47260 |
0.11 |
| chr4_106778453_106778616 | 0.32 |
Gm12745 |
predicted gene 12745 |
3718 |
0.18 |
| chr2_45261991_45262163 | 0.32 |
Gm13475 |
predicted gene 13475 |
67155 |
0.12 |
| chr4_134067460_134067623 | 0.32 |
Crybg2 |
crystallin beta-gamma domain containing 2 |
911 |
0.42 |
| chr17_29123827_29123978 | 0.31 |
Rab44 |
RAB44, member RAS oncogene family |
9721 |
0.09 |
| chr7_48925237_48925540 | 0.31 |
Gm18559 |
predicted gene, 18559 |
14730 |
0.13 |
| chr4_104229316_104229674 | 0.31 |
Dab1 |
disabled 1 |
137618 |
0.05 |
| chr17_5058324_5058580 | 0.31 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
17874 |
0.23 |
| chr17_6197582_6197772 | 0.31 |
Tulp4 |
tubby like protein 4 |
34975 |
0.1 |
| chr10_59537699_59537863 | 0.31 |
Gm10322 |
predicted gene 10322 |
78282 |
0.08 |
| chr8_27106933_27107116 | 0.31 |
Adgra2 |
adhesion G protein-coupled receptor A2 |
7109 |
0.12 |
| chr1_68809984_68810367 | 0.31 |
Gm37061 |
predicted gene, 37061 |
48448 |
0.17 |
| chr11_51584565_51584756 | 0.31 |
Phykpl |
5-phosphohydroxy-L-lysine phospholyase |
97 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.6 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.1 | 0.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.4 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.2 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.0 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) regulation of eosinophil activation(GO:1902566) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.2 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.0 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.0 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:1902992 | negative regulation of amyloid precursor protein catabolic process(GO:1902992) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.2 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.6 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.0 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0035935 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0051088 | monocyte activation(GO:0042117) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.0 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.0 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.0 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.7 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0043813 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |