Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
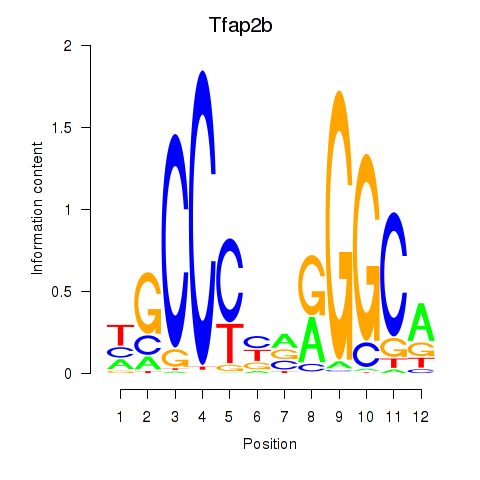
Results for Tfap2b
Z-value: 1.52
Transcription factors associated with Tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2b
|
ENSMUSG00000025927.7 | transcription factor AP-2 beta |
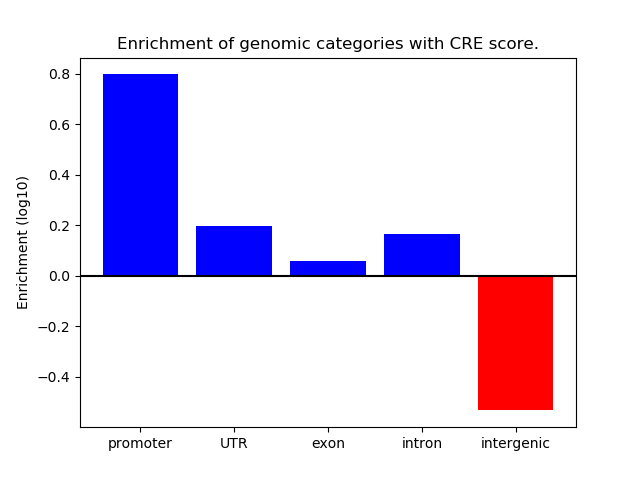
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_19205686_19205886 | Tfap2b | 3128 | 0.260115 | -0.87 | 2.2e-02 | Click! |
Activity of the Tfap2b motif across conditions
Conditions sorted by the z-value of the Tfap2b motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
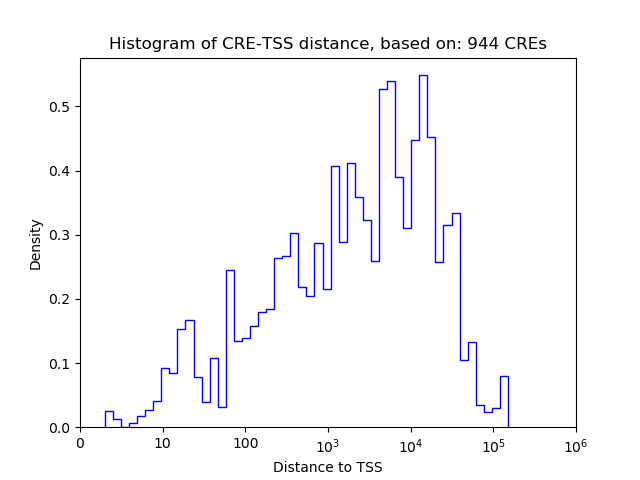
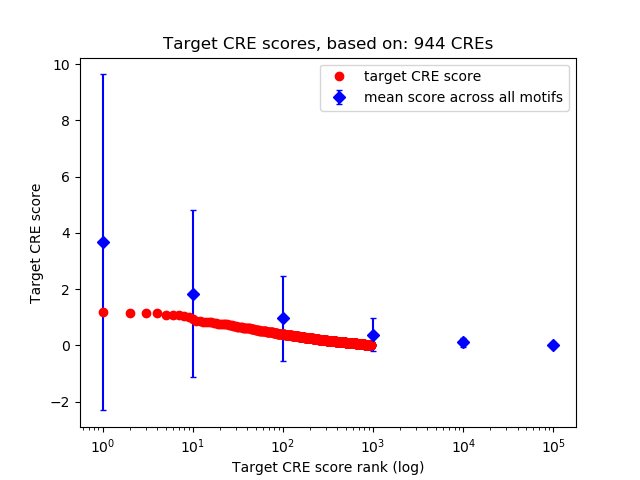
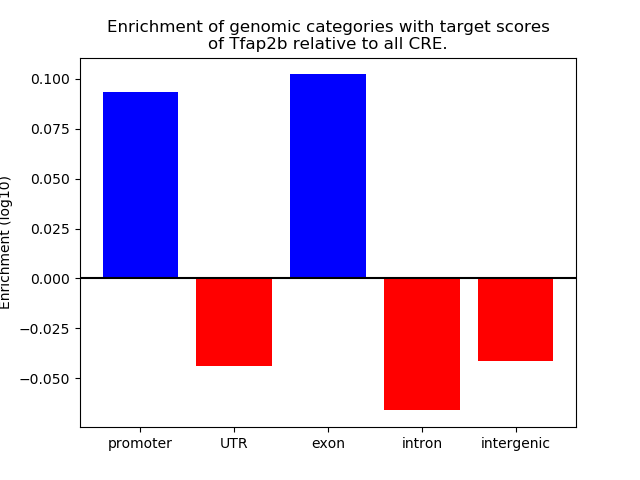
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_45333523_45333723 | 1.19 |
Cyp4v3 |
cytochrome P450, family 4, subfamily v, polypeptide 3 |
407 |
0.81 |
| chr12_108856815_108857551 | 1.15 |
Gm22079 |
predicted gene, 22079 |
5486 |
0.1 |
| chr7_102025342_102025783 | 1.13 |
Rnf121 |
ring finger protein 121 |
1093 |
0.33 |
| chr2_181413297_181413448 | 1.13 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
11016 |
0.1 |
| chr2_167831188_167831379 | 1.09 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
2363 |
0.25 |
| chr1_89669764_89670114 | 1.08 |
4933400F21Rik |
RIKEN cDNA 4933400F21 gene |
13949 |
0.24 |
| chr9_107688467_107688630 | 1.06 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
1231 |
0.26 |
| chr9_50874153_50874482 | 1.06 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
1305 |
0.4 |
| chr10_85829643_85829823 | 1.01 |
Gm19624 |
predicted gene, 19624 |
9 |
0.75 |
| chr4_57320936_57321111 | 0.93 |
Ptpn3 |
protein tyrosine phosphatase, non-receptor type 3 |
19186 |
0.15 |
| chr10_63373532_63373708 | 0.88 |
Sirt1 |
sirtuin 1 |
8084 |
0.13 |
| chr6_128102357_128102567 | 0.86 |
Gm26338 |
predicted gene, 26338 |
7436 |
0.15 |
| chr1_179836517_179836707 | 0.84 |
Ahctf1 |
AT hook containing transcription factor 1 |
32932 |
0.16 |
| chr16_22699066_22699230 | 0.83 |
Gm8118 |
predicted gene 8118 |
12954 |
0.18 |
| chr11_119936702_119936872 | 0.82 |
Gm11766 |
predicted gene 11766 |
1724 |
0.24 |
| chr7_45213806_45213957 | 0.82 |
Dkkl1 |
dickkopf-like 1 |
1734 |
0.12 |
| chr7_143313622_143313928 | 0.81 |
Gm37364 |
predicted gene, 37364 |
16077 |
0.12 |
| chr8_120622299_120622462 | 0.78 |
1190005I06Rik |
RIKEN cDNA 1190005I06 gene |
11983 |
0.09 |
| chr5_110431894_110432052 | 0.77 |
Fbrsl1 |
fibrosin-like 1 |
1348 |
0.35 |
| chr2_179292597_179292961 | 0.75 |
Gm14293 |
predicted gene 14293 |
52289 |
0.14 |
| chr2_69911096_69911271 | 0.75 |
Ubr3 |
ubiquitin protein ligase E3 component n-recognin 3 |
13880 |
0.13 |
| chr10_86685801_86686017 | 0.75 |
1810014B01Rik |
RIKEN cDNA 1810014B01 gene |
20 |
0.93 |
| chr1_133877032_133877215 | 0.75 |
Optc |
opticin |
24204 |
0.12 |
| chr1_167387431_167387591 | 0.75 |
Mgst3 |
microsomal glutathione S-transferase 3 |
6330 |
0.16 |
| chr2_149759276_149759438 | 0.74 |
Gm14130 |
predicted gene 14130 |
16182 |
0.2 |
| chr6_112883814_112883983 | 0.73 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
54439 |
0.1 |
| chr11_55406169_55406386 | 0.71 |
Sparc |
secreted acidic cysteine rich glycoprotein |
4112 |
0.19 |
| chr7_19228252_19228443 | 0.70 |
Opa3 |
optic atrophy 3 |
13 |
0.94 |
| chr12_76710114_76710283 | 0.68 |
Sptb |
spectrin beta, erythrocytic |
175 |
0.95 |
| chr7_28969689_28969854 | 0.67 |
Eif3k |
eukaryotic translation initiation factor 3, subunit K |
5065 |
0.11 |
| chr16_4912876_4913042 | 0.67 |
Mgrn1 |
mahogunin, ring finger 1 |
252 |
0.86 |
| chr9_43467006_43467175 | 0.67 |
Gm28215 |
predicted gene 28215 |
2332 |
0.29 |
| chr11_101369464_101369615 | 0.65 |
G6pc |
glucose-6-phosphatase, catalytic |
1978 |
0.11 |
| chr1_165629214_165629365 | 0.65 |
Mpzl1 |
myelin protein zero-like 1 |
4797 |
0.13 |
| chr1_153990768_153990949 | 0.64 |
Gm29291 |
predicted gene 29291 |
15359 |
0.15 |
| chr4_148448716_148449227 | 0.63 |
Mtor |
mechanistic target of rapamycin kinase |
346 |
0.82 |
| chr19_3850863_3851029 | 0.62 |
Chka |
choline kinase alpha |
827 |
0.37 |
| chr10_39174337_39174497 | 0.62 |
D030034A15Rik |
RIKEN cDNA D030034A15 gene |
1165 |
0.4 |
| chr7_49178293_49178474 | 0.62 |
Gm37613 |
predicted gene, 37613 |
17158 |
0.18 |
| chr17_29394251_29394433 | 0.62 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
17841 |
0.11 |
| chr14_51100561_51100914 | 0.61 |
Rnase4 |
ribonuclease, RNase A family 4 |
4378 |
0.09 |
| chr10_96377652_96377803 | 0.61 |
Gm20091 |
predicted gene, 20091 |
31311 |
0.14 |
| chr13_32066979_32067135 | 0.60 |
Gm48885 |
predicted gene, 48885 |
32118 |
0.21 |
| chr11_50292381_50292544 | 0.59 |
Maml1 |
mastermind like transcriptional coactivator 1 |
151 |
0.93 |
| chr8_120437914_120438125 | 0.58 |
Gm22715 |
predicted gene, 22715 |
5530 |
0.21 |
| chr5_104399237_104399791 | 0.58 |
Spp1 |
secreted phosphoprotein 1 |
35604 |
0.12 |
| chr1_165461123_165461290 | 0.57 |
Mpc2 |
mitochondrial pyruvate carrier 2 |
18 |
0.92 |
| chr9_59539398_59539715 | 0.56 |
Hexa |
hexosaminidase A |
16 |
0.8 |
| chr5_139818915_139819083 | 0.56 |
Tmem184a |
transmembrane protein 184a |
918 |
0.45 |
| chr18_31910656_31910818 | 0.55 |
Sft2d3 |
SFT2 domain containing 3 |
1087 |
0.41 |
| chr17_32241257_32241435 | 0.55 |
Gm4432 |
predicted gene 4432 |
817 |
0.53 |
| chr16_22163049_22163219 | 0.54 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
115 |
0.96 |
| chr11_45877769_45877933 | 0.54 |
Clint1 |
clathrin interactor 1 |
4546 |
0.19 |
| chr5_135187024_135187335 | 0.53 |
Baz1b |
bromodomain adjacent to zinc finger domain, 1B |
85 |
0.95 |
| chr12_111121851_111122115 | 0.52 |
Gm48632 |
predicted gene, 48632 |
30783 |
0.12 |
| chr15_102243536_102243699 | 0.52 |
Rarg |
retinoic acid receptor, gamma |
28 |
0.95 |
| chr8_119419822_119419973 | 0.51 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
14227 |
0.14 |
| chr13_54249070_54249391 | 0.51 |
Gm48615 |
predicted gene, 48615 |
3934 |
0.19 |
| chr6_125088559_125088736 | 0.51 |
2010008C14Rik |
RIKEN cDNA 2010008C14 gene |
5210 |
0.08 |
| chr2_124858722_124858893 | 0.51 |
Gm13994 |
predicted gene 13994 |
150152 |
0.04 |
| chr1_191946290_191946441 | 0.50 |
Rd3 |
retinal degeneration 3 |
31005 |
0.1 |
| chr16_91822408_91822590 | 0.50 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
5038 |
0.2 |
| chr10_75596123_75596274 | 0.50 |
Gm9985 |
predicted gene 9985 |
5545 |
0.11 |
| chr12_85151515_85151693 | 0.50 |
Rps6kl1 |
ribosomal protein S6 kinase-like 1 |
340 |
0.8 |
| chr10_127659568_127659719 | 0.49 |
Stat6 |
signal transducer and activator of transcription 6 |
243 |
0.82 |
| chr5_43032061_43032271 | 0.49 |
Gm43700 |
predicted gene 43700 |
6070 |
0.19 |
| chr7_19572974_19573173 | 0.49 |
Gemin7 |
gem nuclear organelle associated protein 7 |
16 |
0.95 |
| chr7_18957606_18957786 | 0.48 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
29704 |
0.06 |
| chr8_128491169_128491342 | 0.48 |
Nrp1 |
neuropilin 1 |
131858 |
0.05 |
| chr11_100515071_100515366 | 0.48 |
Acly |
ATP citrate lyase |
8380 |
0.1 |
| chr6_43292582_43293012 | 0.48 |
Arhgef5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
9915 |
0.12 |
| chr4_118247695_118247892 | 0.47 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
11348 |
0.17 |
| chr10_75121130_75121291 | 0.47 |
Bcr |
BCR activator of RhoGEF and GTPase |
39150 |
0.15 |
| chr16_30525931_30526082 | 0.47 |
Tmem44 |
transmembrane protein 44 |
14903 |
0.19 |
| chr7_30347923_30348110 | 0.46 |
4930479H17Rik |
RIKEN cDNA 4930479H17 gene |
5305 |
0.07 |
| chr6_118593826_118593977 | 0.46 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1525 |
0.4 |
| chr19_9954439_9954606 | 0.46 |
Gm50346 |
predicted gene, 50346 |
11690 |
0.09 |
| chr3_81036630_81036817 | 0.45 |
Pdgfc |
platelet-derived growth factor, C polypeptide |
307 |
0.87 |
| chr2_165969012_165969173 | 0.44 |
Gm11462 |
predicted gene 11462 |
9303 |
0.13 |
| chr14_14345872_14346046 | 0.44 |
Il3ra |
interleukin 3 receptor, alpha chain |
316 |
0.78 |
| chr13_95554131_95554549 | 0.44 |
Gm48730 |
predicted gene, 48730 |
18750 |
0.14 |
| chr4_134061289_134061592 | 0.43 |
Crybg2 |
crystallin beta-gamma domain containing 2 |
625 |
0.58 |
| chr10_126979601_126979791 | 0.43 |
Ctdsp2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
667 |
0.48 |
| chr10_77144461_77144617 | 0.43 |
Gm7775 |
predicted gene 7775 |
7439 |
0.19 |
| chr8_120538960_120539167 | 0.43 |
Mir7687 |
microRNA 7687 |
367 |
0.7 |
| chr7_113242447_113242773 | 0.42 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
2856 |
0.29 |
| chr5_112283643_112283806 | 0.42 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
5038 |
0.13 |
| chr18_80236503_80236751 | 0.41 |
Hsbp1l1 |
heat shock factor binding protein 1-like 1 |
206 |
0.89 |
| chr14_70520498_70520830 | 0.41 |
Bmp1 |
bone morphogenetic protein 1 |
430 |
0.67 |
| chr9_54516094_54516256 | 0.41 |
Dmxl2 |
Dmx-like 2 |
14549 |
0.17 |
| chr7_44858584_44858824 | 0.41 |
Pnkp |
polynucleotide kinase 3'- phosphatase |
208 |
0.82 |
| chr2_11365489_11365685 | 0.41 |
Gm37730 |
predicted gene, 37730 |
1864 |
0.2 |
| chr6_128803059_128803375 | 0.41 |
Klrb1c |
killer cell lectin-like receptor subfamily B member 1C |
14002 |
0.09 |
| chr19_46003413_46003585 | 0.41 |
Hps6 |
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
9 |
0.96 |
| chr12_80001998_80002169 | 0.40 |
Gm8275 |
predicted gene 8275 |
22232 |
0.17 |
| chr2_155099563_155099720 | 0.40 |
2310005A03Rik |
RIKEN cDNA 2310005A03 gene |
440 |
0.77 |
| chr6_29272524_29272714 | 0.40 |
Hilpda |
hypoxia inducible lipid droplet associated |
7 |
0.96 |
| chr3_101599885_101600061 | 0.39 |
Gm42941 |
predicted gene 42941 |
2843 |
0.23 |
| chr2_167842569_167842745 | 0.39 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
9011 |
0.16 |
| chr7_46835248_46835408 | 0.39 |
Gm45308 |
predicted gene 45308 |
2864 |
0.13 |
| chr2_119742132_119742303 | 0.39 |
Itpka |
inositol 1,4,5-trisphosphate 3-kinase A |
120 |
0.91 |
| chr9_90113472_90114008 | 0.39 |
Morf4l1 |
mortality factor 4 like 1 |
549 |
0.72 |
| chr11_118211857_118212019 | 0.39 |
Cyth1 |
cytohesin 1 |
804 |
0.57 |
| chr17_43327081_43327253 | 0.39 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
33284 |
0.19 |
| chr14_30011884_30012221 | 0.38 |
Chdh |
choline dehydrogenase |
2881 |
0.15 |
| chr11_96863519_96863691 | 0.38 |
Gm11524 |
predicted gene 11524 |
6590 |
0.09 |
| chr11_106491531_106491682 | 0.38 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
3754 |
0.18 |
| chr4_106980690_106980853 | 0.38 |
Ssbp3 |
single-stranded DNA binding protein 3 |
14345 |
0.18 |
| chr19_3877298_3877449 | 0.38 |
Chka |
choline kinase alpha |
1968 |
0.16 |
| chr4_150065927_150066095 | 0.38 |
Mir34a |
microRNA 34a |
2443 |
0.16 |
| chr4_123299380_123299675 | 0.38 |
Pabpc4 |
poly(A) binding protein, cytoplasmic 4 |
2347 |
0.15 |
| chr5_105826898_105827072 | 0.38 |
Lrrc8d |
leucine rich repeat containing 8D |
2474 |
0.21 |
| chr11_101928490_101928696 | 0.38 |
Rpl27-ps2 |
ribosomal protein L27, pseudogene 2 |
10908 |
0.1 |
| chr11_44405875_44406089 | 0.38 |
Il12b |
interleukin 12b |
5919 |
0.19 |
| chr7_99699741_99699908 | 0.37 |
Slco2b1 |
solute carrier organic anion transporter family, member 2b1 |
4015 |
0.12 |
| chr8_119445523_119446899 | 0.37 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
508 |
0.74 |
| chr1_86586776_86586949 | 0.37 |
Cops7b |
COP9 signalosome subunit 7B |
238 |
0.88 |
| chr19_42551158_42551329 | 0.36 |
Gm16541 |
predicted gene 16541 |
11852 |
0.18 |
| chr14_61360115_61360286 | 0.36 |
Ebpl |
emopamil binding protein-like |
120 |
0.94 |
| chr8_124275783_124275948 | 0.36 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
19238 |
0.16 |
| chr1_88959907_88960074 | 0.36 |
Gm4753 |
predicted gene 4753 |
27671 |
0.17 |
| chr4_137769010_137769180 | 0.36 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
520 |
0.8 |
| chr10_84757574_84757782 | 0.35 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
1616 |
0.43 |
| chr5_110797818_110798001 | 0.35 |
Ulk1 |
unc-51 like kinase 1 |
2075 |
0.19 |
| chr16_32246842_32247035 | 0.35 |
Fbxo45 |
F-box protein 45 |
220 |
0.55 |
| chr17_17345622_17345809 | 0.35 |
Oaz1-ps |
ornithine decarboxylase antizyme 1, pseudogene |
425 |
0.8 |
| chr9_31279892_31280066 | 0.35 |
Prdm10 |
PR domain containing 10 |
559 |
0.59 |
| chr4_137824947_137825262 | 0.34 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
28720 |
0.16 |
| chr9_40628453_40628756 | 0.34 |
Gm48284 |
predicted gene, 48284 |
12218 |
0.12 |
| chr7_110942267_110942426 | 0.34 |
Mrvi1 |
MRV integration site 1 |
3841 |
0.23 |
| chr10_40124800_40124982 | 0.34 |
Gm25613 |
predicted gene, 25613 |
15429 |
0.13 |
| chr12_108832369_108832526 | 0.34 |
Slc25a29 |
solute carrier family 25 (mitochondrial carrier, palmitoylcarnitine transporter), member 29 |
830 |
0.42 |
| chr6_54070751_54070926 | 0.34 |
Chn2 |
chimerin 2 |
26882 |
0.18 |
| chr15_83518459_83518650 | 0.34 |
Gm24575 |
predicted gene, 24575 |
1618 |
0.25 |
| chr9_57785755_57786023 | 0.34 |
Arid3b |
AT rich interactive domain 3B (BRIGHT-like) |
5759 |
0.16 |
| chr1_93370316_93370486 | 0.34 |
Ano7 |
anoctamin 7 |
3529 |
0.12 |
| chr7_140835414_140835584 | 0.33 |
Urah |
urate (5-hydroxyiso-) hydrolase |
125 |
0.89 |
| chr16_29944225_29944722 | 0.33 |
Gm26569 |
predicted gene, 26569 |
2043 |
0.31 |
| chr16_78377023_78377174 | 0.33 |
Btg3 |
BTG anti-proliferation factor 3 |
94 |
0.97 |
| chr8_70221071_70221246 | 0.33 |
Armc6 |
armadillo repeat containing 6 |
1132 |
0.32 |
| chr7_83636527_83636716 | 0.33 |
Gm45838 |
predicted gene 45838 |
3250 |
0.14 |
| chr2_132721307_132721479 | 0.33 |
Gm22245 |
predicted gene, 22245 |
4287 |
0.14 |
| chr2_167634494_167634653 | 0.33 |
Ube2v1 |
ubiquitin-conjugating enzyme E2 variant 1 |
2478 |
0.16 |
| chr3_85887151_85887500 | 0.32 |
Gm37240 |
predicted gene, 37240 |
43 |
0.62 |
| chr6_31399464_31399657 | 0.32 |
Mkln1 |
muskelin 1, intracellular mediator containing kelch motifs |
701 |
0.44 |
| chr2_119355713_119355881 | 0.32 |
Chac1 |
ChaC, cation transport regulator 1 |
4568 |
0.16 |
| chr11_120189789_120189958 | 0.32 |
2810410L24Rik |
RIKEN cDNA 2810410L24 gene |
21 |
0.96 |
| chr7_141818371_141818522 | 0.32 |
Muc5b |
mucin 5, subtype B, tracheobronchial |
20624 |
0.14 |
| chr11_97819881_97820206 | 0.31 |
Lasp1 |
LIM and SH3 protein 1 |
4795 |
0.1 |
| chr6_99521816_99522204 | 0.31 |
Foxp1 |
forkhead box P1 |
637 |
0.76 |
| chr9_106447373_106447582 | 0.31 |
Abhd14b |
abhydrolase domain containing 14b |
18 |
0.49 |
| chr3_94690762_94690922 | 0.31 |
Selenbp2 |
selenium binding protein 2 |
2714 |
0.16 |
| chr5_92110928_92111278 | 0.31 |
Gm24931 |
predicted gene, 24931 |
7180 |
0.13 |
| chr4_3938729_3938917 | 0.31 |
Chchd7 |
coiled-coil-helix-coiled-coil-helix domain containing 7 |
65 |
0.84 |
| chr15_86169783_86169988 | 0.31 |
Cerk |
ceramide kinase |
10417 |
0.17 |
| chr13_31223115_31223275 | 0.30 |
4930401O12Rik |
RIKEN cDNA 4930401O12 gene |
17891 |
0.19 |
| chr1_165768492_165768643 | 0.30 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
909 |
0.37 |
| chr6_121879272_121879452 | 0.30 |
Mug1 |
murinoglobulin 1 |
6215 |
0.21 |
| chr18_80704875_80705052 | 0.30 |
Gm50211 |
predicted gene, 50211 |
227 |
0.92 |
| chr1_61649997_61650160 | 0.30 |
Gm37205 |
predicted gene, 37205 |
4345 |
0.17 |
| chr3_10284943_10285269 | 0.30 |
Fabp12 |
fatty acid binding protein 12 |
16068 |
0.09 |
| chr1_119648939_119649109 | 0.30 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
24 |
0.97 |
| chrX_159988087_159988260 | 0.30 |
Map3k15 |
mitogen-activated protein kinase kinase kinase 15 |
260 |
0.92 |
| chr8_84656944_84657116 | 0.29 |
Ier2 |
immediate early response 2 |
5824 |
0.11 |
| chr16_11006083_11006392 | 0.29 |
Gm24961 |
predicted gene, 24961 |
11851 |
0.1 |
| chr16_95679759_95679918 | 0.29 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
22237 |
0.2 |
| chr1_87803823_87803974 | 0.29 |
Sag |
S-antigen, retina and pineal gland (arrestin) |
218 |
0.9 |
| chr5_51511559_51511740 | 0.29 |
Gm43605 |
predicted gene 43605 |
2644 |
0.23 |
| chr9_57650943_57651123 | 0.29 |
Csk |
c-src tyrosine kinase |
2438 |
0.18 |
| chr6_124712989_124713179 | 0.29 |
Phb2 |
prohibitin 2 |
38 |
0.88 |
| chr2_103713943_103714113 | 0.29 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
1007 |
0.49 |
| chr5_139364533_139364950 | 0.28 |
Mir339 |
microRNA 339 |
5004 |
0.12 |
| chr4_63252131_63252305 | 0.28 |
Mir455 |
microRNA 455 |
4633 |
0.19 |
| chr2_72179596_72179778 | 0.28 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
6 |
0.98 |
| chr14_47472459_47472619 | 0.28 |
Fbxo34 |
F-box protein 34 |
22 |
0.96 |
| chr2_167834679_167834867 | 0.27 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
1127 |
0.46 |
| chr13_34742265_34742434 | 0.27 |
Fam50b |
family with sequence similarity 50, member B |
2737 |
0.19 |
| chr13_58960766_58960939 | 0.27 |
Ntrk2 |
neurotrophic tyrosine kinase, receptor, type 2 |
89117 |
0.07 |
| chr7_19797262_19797473 | 0.27 |
Cblc |
Casitas B-lineage lymphoma c |
558 |
0.52 |
| chr9_21546507_21546700 | 0.27 |
Carm1 |
coactivator-associated arginine methyltransferase 1 |
291 |
0.82 |
| chrX_20425191_20425364 | 0.27 |
Jade3 |
jade family PHD finger 3 |
411 |
0.81 |
| chr4_53440081_53440358 | 0.27 |
Slc44a1 |
solute carrier family 44, member 1 |
194 |
0.96 |
| chr7_127886041_127886230 | 0.27 |
Prss53 |
protease, serine 53 |
4832 |
0.07 |
| chr11_5846893_5847128 | 0.27 |
Polm |
polymerase (DNA directed), mu |
8994 |
0.11 |
| chr9_103007772_103008092 | 0.26 |
Slco2a1 |
solute carrier organic anion transporter family, member 2a1 |
70 |
0.97 |
| chr9_117357124_117357284 | 0.26 |
Gm20396 |
predicted gene 20396 |
45775 |
0.18 |
| chr4_133967614_133967768 | 0.26 |
Hmgn2 |
high mobility group nucleosomal binding domain 2 |
170 |
0.93 |
| chr7_75762457_75762611 | 0.26 |
Gm23251 |
predicted gene, 23251 |
5166 |
0.17 |
| chr7_133728557_133728948 | 0.26 |
Dhx32 |
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
2148 |
0.2 |
| chr2_5669776_5669983 | 0.26 |
Camk1d |
calcium/calmodulin-dependent protein kinase ID |
6167 |
0.27 |
| chr8_83738255_83738433 | 0.26 |
Adgre5 |
adhesion G protein-coupled receptor E5 |
2825 |
0.16 |
| chr13_46888106_46888269 | 0.26 |
Gm48250 |
predicted gene, 48250 |
7160 |
0.15 |
| chr9_71153822_71154046 | 0.26 |
Aqp9 |
aquaporin 9 |
8699 |
0.18 |
| chr8_94186443_94186638 | 0.26 |
Gm39228 |
predicted gene, 39228 |
3251 |
0.13 |
| chr15_82378546_82378931 | 0.26 |
Cyp2d22 |
cytochrome P450, family 2, subfamily d, polypeptide 22 |
1505 |
0.16 |
| chr8_116581400_116581581 | 0.26 |
Dynlrb2 |
dynein light chain roadblock-type 2 |
76475 |
0.1 |
| chr15_90679373_90679524 | 0.25 |
Cpne8 |
copine VIII |
16 |
0.98 |
| chr18_11205176_11205343 | 0.25 |
Gata6 |
GATA binding protein 6 |
146212 |
0.04 |
| chr11_109487198_109487366 | 0.25 |
Arsg |
arylsulfatase G |
1676 |
0.26 |
| chr2_168003768_168003979 | 0.25 |
Gm14236 |
predicted gene 14236 |
6656 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 0.5 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.2 | 0.5 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.4 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 0.3 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.1 | 0.4 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.2 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.1 | 0.2 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.1 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.3 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.3 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.5 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.3 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:1902661 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.2 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.1 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0032353 | negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) |
| 0.0 | 0.0 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0072300 | positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.0 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.0 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.7 | GO:0034560 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.2 | GO:0034943 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |