Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
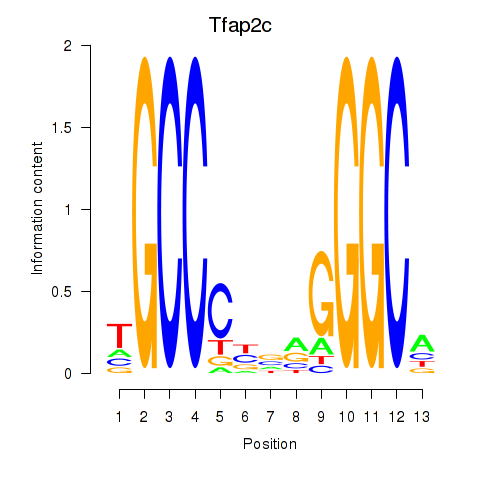
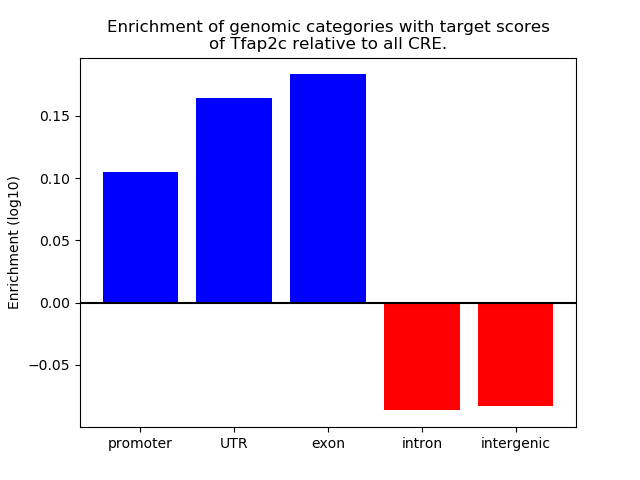
Results for Tfap2c
Z-value: 1.29
Transcription factors associated with Tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2c
|
ENSMUSG00000028640.5 | transcription factor AP-2, gamma |
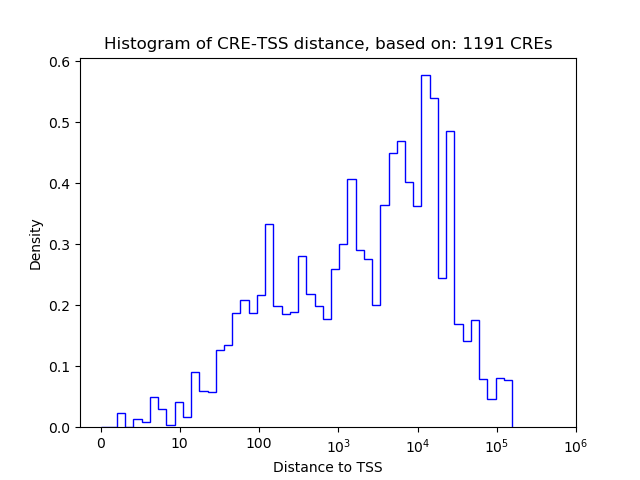
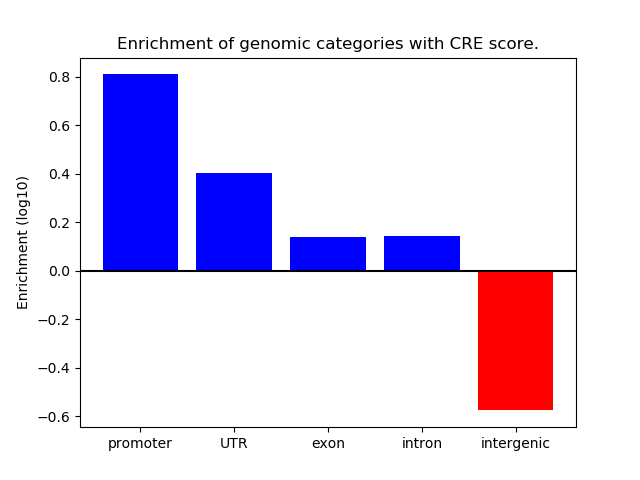
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_172684895_172685060 | Tfap2c | 132861 | 0.043166 | 0.72 | 1.0e-01 | Click! |
| chr2_172550558_172550733 | Tfap2c | 8 | 0.979771 | -0.71 | 1.2e-01 | Click! |
| chr2_172614148_172614317 | Tfap2c | 62116 | 0.114609 | -0.63 | 1.8e-01 | Click! |
| chr2_172580659_172580839 | Tfap2c | 28633 | 0.180876 | 0.52 | 2.9e-01 | Click! |
| chr2_172690322_172690473 | Tfap2c | 138281 | 0.040437 | 0.45 | 3.7e-01 | Click! |
Activity of the Tfap2c motif across conditions
Conditions sorted by the z-value of the Tfap2c motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
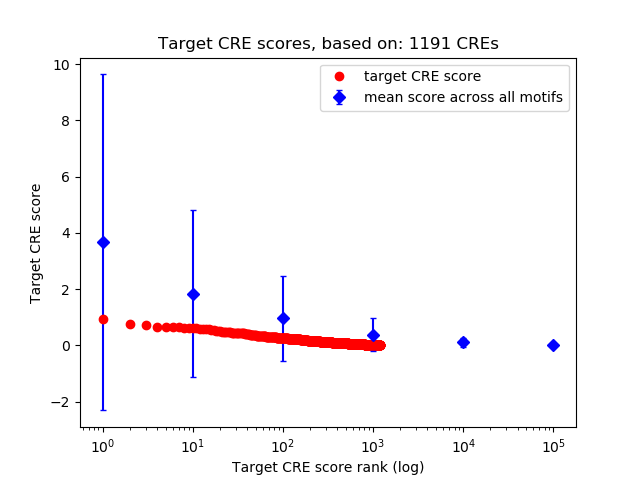
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_99104837_99105019 | 0.93 |
Tns4 |
tensin 4 |
15622 |
0.14 |
| chr4_138979154_138979535 | 0.76 |
Tmco4 |
transmembrane and coiled-coil domains 4 |
6435 |
0.16 |
| chr10_80438140_80438330 | 0.72 |
Tcf3 |
transcription factor 3 |
4588 |
0.1 |
| chr7_19952685_19952933 | 0.67 |
Rpl7a-ps8 |
ribosomal protein L7A, pseudogene 8 |
974 |
0.29 |
| chr11_52257389_52257568 | 0.67 |
Tcf7 |
transcription factor 7, T cell specific |
17399 |
0.11 |
| chr7_79279429_79279594 | 0.67 |
Gm44639 |
predicted gene 44639 |
1744 |
0.27 |
| chr5_99239712_99239874 | 0.64 |
Rasgef1b |
RasGEF domain family, member 1B |
3447 |
0.29 |
| chr13_28930434_28930607 | 0.62 |
Sox4 |
SRY (sex determining region Y)-box 4 |
23193 |
0.17 |
| chr5_144370961_144371328 | 0.61 |
Dmrt1i |
Dmrt1 interacting ncRNA |
12619 |
0.16 |
| chr18_65989569_65989860 | 0.60 |
Lman1 |
lectin, mannose-binding, 1 |
3946 |
0.15 |
| chr14_11619733_11619902 | 0.60 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
66236 |
0.11 |
| chr16_8659432_8659599 | 0.59 |
Pmm2 |
phosphomannomutase 2 |
10923 |
0.1 |
| chr4_57543982_57544152 | 0.59 |
Pakap |
paralemmin A kinase anchor protein |
24092 |
0.22 |
| chr5_122459012_122459200 | 0.58 |
Anapc7 |
anaphase promoting complex subunit 7 |
17731 |
0.08 |
| chr16_13940553_13940716 | 0.57 |
Mpv17l |
Mpv17 transgene, kidney disease mutant-like |
4 |
0.96 |
| chr1_169166546_169166738 | 0.54 |
Mir6354 |
microRNA 6354 |
147553 |
0.04 |
| chr10_78308725_78308893 | 0.54 |
AC160405.1 |
|
1190 |
0.23 |
| chr17_63488193_63488355 | 0.50 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
4479 |
0.29 |
| chr17_56780401_56780582 | 0.50 |
Rfx2 |
regulatory factor X, 2 (influences HLA class II expression) |
3213 |
0.14 |
| chr5_151060423_151060640 | 0.49 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
28731 |
0.19 |
| chr7_46726457_46726608 | 0.48 |
Saa4 |
serum amyloid A 4 |
6071 |
0.09 |
| chr16_46010174_46010340 | 0.48 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
39 |
0.97 |
| chr8_72409462_72409669 | 0.48 |
Eps15l1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
11873 |
0.1 |
| chr10_80332281_80332447 | 0.47 |
Reep6 |
receptor accessory protein 6 |
2162 |
0.11 |
| chr16_24313632_24313811 | 0.46 |
AC169509.1 |
BMP2 inducible kinase (Bmp2k) pseudogene |
23885 |
0.17 |
| chr2_26153414_26153565 | 0.46 |
Tmem250-ps |
transmembrane protein 250, pseudogene |
12968 |
0.11 |
| chr11_87732236_87732387 | 0.45 |
Rnf43 |
ring finger protein 43 |
1609 |
0.21 |
| chr16_97940453_97940821 | 0.45 |
Zbtb21 |
zinc finger and BTB domain containing 21 |
12461 |
0.17 |
| chr4_133561732_133562055 | 0.45 |
Gm23158 |
predicted gene, 23158 |
2944 |
0.13 |
| chr18_67448711_67448888 | 0.44 |
Afg3l2 |
AFG3-like AAA ATPase 2 |
308 |
0.86 |
| chr19_8987926_8988110 | 0.44 |
Ahnak |
AHNAK nucleoprotein (desmoyokin) |
1266 |
0.26 |
| chr8_40660395_40660561 | 0.44 |
Adam24 |
a disintegrin and metallopeptidase domain 24 (testase 1) |
14599 |
0.14 |
| chr3_104707896_104708056 | 0.44 |
Gm29572 |
predicted gene 29572 |
1391 |
0.27 |
| chr2_172511032_172511203 | 0.43 |
Gm14303 |
predicted gene 14303 |
1426 |
0.37 |
| chr2_27042057_27042419 | 0.42 |
Gm13398 |
predicted gene 13398 |
4953 |
0.11 |
| chr10_84872038_84872201 | 0.42 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
33971 |
0.16 |
| chr18_45575250_45575614 | 0.42 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
812 |
0.69 |
| chr1_74807002_74807292 | 0.42 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
13784 |
0.11 |
| chr5_33657811_33658158 | 0.41 |
Tmem129 |
transmembrane protein 129 |
85 |
0.57 |
| chr19_55463767_55463937 | 0.41 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
82838 |
0.1 |
| chr7_46849448_46849605 | 0.41 |
Ldha |
lactate dehydrogenase A |
539 |
0.6 |
| chr7_145047841_145048023 | 0.40 |
Ccnd1 |
cyclin D1 |
108007 |
0.05 |
| chr19_10596220_10596418 | 0.37 |
Tkfc |
triokinase, FMN cyclase |
7939 |
0.09 |
| chr5_34527351_34527633 | 0.37 |
Sh3bp2 |
SH3-domain binding protein 2 |
1646 |
0.22 |
| chr9_108400984_108401135 | 0.37 |
1700102P08Rik |
RIKEN cDNA 1700102P08 gene |
8225 |
0.07 |
| chr7_143614809_143614986 | 0.37 |
Tnfrsf26 |
tumor necrosis factor receptor superfamily, member 26 |
13056 |
0.09 |
| chr11_8629102_8629275 | 0.36 |
Tns3 |
tensin 3 |
4301 |
0.35 |
| chr11_35930971_35931124 | 0.36 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
49480 |
0.14 |
| chr4_131924003_131924348 | 0.36 |
Tmem200b |
transmembrane protein 200B |
2404 |
0.15 |
| chr7_99168829_99169197 | 0.36 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
1093 |
0.4 |
| chr11_103689147_103689298 | 0.35 |
Gosr2 |
golgi SNAP receptor complex member 2 |
8472 |
0.13 |
| chr5_30523230_30523387 | 0.35 |
Gm42765 |
predicted gene 42765 |
6149 |
0.14 |
| chr12_100996285_100996501 | 0.34 |
Gm36756 |
predicted gene, 36756 |
3421 |
0.16 |
| chr6_86263585_86263762 | 0.34 |
Gm43917 |
predicted gene, 43917 |
241 |
0.9 |
| chr19_10065348_10065653 | 0.34 |
Fads2 |
fatty acid desaturase 2 |
5687 |
0.14 |
| chr3_104670885_104671048 | 0.33 |
Gm29560 |
predicted gene 29560 |
956 |
0.37 |
| chr5_118664076_118664234 | 0.33 |
Gm43274 |
predicted gene 43274 |
19782 |
0.14 |
| chr4_152232556_152232715 | 0.33 |
Gm13096 |
predicted gene 13096 |
28709 |
0.1 |
| chr12_104992554_104993025 | 0.33 |
Syne3 |
spectrin repeat containing, nuclear envelope family member 3 |
5888 |
0.13 |
| chr2_163481751_163481907 | 0.32 |
Fitm2 |
fat storage-inducing transmembrane protein 2 |
9200 |
0.11 |
| chr5_25019324_25019486 | 0.32 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
23652 |
0.16 |
| chr6_90772172_90772486 | 0.32 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
8188 |
0.18 |
| chr1_95317084_95317247 | 0.32 |
Fam174a |
family with sequence similarity 174, member A |
3537 |
0.27 |
| chr4_152062432_152062592 | 0.32 |
Plekhg5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
9986 |
0.11 |
| chr2_166100923_166101091 | 0.31 |
Sulf2 |
sulfatase 2 |
22888 |
0.16 |
| chr16_34110699_34110875 | 0.31 |
Kalrn |
kalirin, RhoGEF kinase |
13697 |
0.26 |
| chr6_139588514_139588690 | 0.31 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
1365 |
0.44 |
| chr2_148022052_148022231 | 0.31 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
16129 |
0.17 |
| chr9_108765050_108765207 | 0.31 |
Prkar2a |
protein kinase, cAMP dependent regulatory, type II alpha |
16883 |
0.09 |
| chr10_81320539_81320949 | 0.31 |
Cactin |
cactin, spliceosome C complex subunit |
359 |
0.64 |
| chr13_55450624_55450785 | 0.30 |
Mir6943 |
microRNA 6943 |
2402 |
0.13 |
| chr11_69415319_69415489 | 0.30 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
1729 |
0.17 |
| chr9_65391751_65391920 | 0.30 |
Kbtbd13 |
kelch repeat and BTB (POZ) domain containing 13 |
183 |
0.89 |
| chr9_9188372_9188561 | 0.30 |
Gm16833 |
predicted gene, 16833 |
47822 |
0.15 |
| chr11_102401993_102402497 | 0.30 |
Slc25a39 |
solute carrier family 25, member 39 |
2248 |
0.14 |
| chr1_191026198_191026354 | 0.30 |
Flvcr1 |
feline leukemia virus subgroup C cellular receptor 1 |
118 |
0.89 |
| chr17_29046089_29046300 | 0.29 |
Srsf3 |
serine and arginine-rich splicing factor 3 |
6500 |
0.09 |
| chr2_18957058_18957209 | 0.29 |
Pip4k2a |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
40993 |
0.15 |
| chr18_37997513_37997702 | 0.29 |
Arap3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
33 |
0.95 |
| chr7_126280513_126280705 | 0.28 |
Sbk1 |
SH3-binding kinase 1 |
7209 |
0.12 |
| chr8_119445523_119446899 | 0.28 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
508 |
0.74 |
| chr18_80261215_80262161 | 0.28 |
Slc66a2 |
solute carrier family 66 member 2 |
1152 |
0.35 |
| chr8_125337571_125337768 | 0.28 |
Gm16237 |
predicted gene 16237 |
110346 |
0.06 |
| chr1_134955895_134956232 | 0.28 |
Ppp1r12b |
protein phosphatase 1, regulatory subunit 12B |
121 |
0.96 |
| chr5_32092059_32092277 | 0.28 |
Babam2 |
BRISC and BRCA1 A complex member 2 |
12471 |
0.17 |
| chr16_22895393_22895557 | 0.28 |
Ahsg |
alpha-2-HS-glycoprotein |
611 |
0.52 |
| chr18_38494979_38495150 | 0.28 |
Gm50349 |
predicted gene, 50349 |
13761 |
0.17 |
| chr12_104863757_104863919 | 0.27 |
Clmn |
calmin |
1171 |
0.39 |
| chr1_178600841_178601326 | 0.27 |
Gm24405 |
predicted gene, 24405 |
35822 |
0.18 |
| chr1_38127167_38127331 | 0.27 |
Rev1 |
REV1, DNA directed polymerase |
1052 |
0.49 |
| chr1_90967653_90967817 | 0.27 |
Rab17 |
RAB17, member RAS oncogene family |
68 |
0.96 |
| chr10_121486376_121486686 | 0.27 |
Gm40787 |
predicted gene, 40787 |
2455 |
0.18 |
| chr7_71216875_71217198 | 0.26 |
6030442E23Rik |
RIKEN cDNA 6030442E23 gene |
13 |
0.98 |
| chr10_93641039_93641238 | 0.26 |
Ntn4 |
netrin 4 |
89 |
0.96 |
| chr1_194482432_194482667 | 0.26 |
Plxna2 |
plexin A2 |
135669 |
0.05 |
| chr12_103995803_103995968 | 0.26 |
Gm28577 |
predicted gene 28577 |
694 |
0.52 |
| chr4_136172527_136172687 | 0.26 |
E2f2 |
E2F transcription factor 2 |
213 |
0.91 |
| chr15_73791838_73792025 | 0.26 |
Ndufb4c |
NADH:ubiquinone oxidoreductase subunit B4C |
17965 |
0.15 |
| chr5_65435787_65435980 | 0.26 |
Ugdh |
UDP-glucose dehydrogenase |
66 |
0.94 |
| chr10_127576673_127576991 | 0.26 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
5053 |
0.12 |
| chr7_49351868_49352264 | 0.26 |
Gm44913 |
predicted gene 44913 |
5972 |
0.24 |
| chr8_11005778_11005929 | 0.26 |
9530052E02Rik |
RIKEN cDNA 9530052E02 gene |
1997 |
0.22 |
| chr13_73767873_73768041 | 0.25 |
Slc12a7 |
solute carrier family 12, member 7 |
4218 |
0.2 |
| chr5_145222977_145223149 | 0.25 |
Zfp655 |
zinc finger protein 655 |
8652 |
0.1 |
| chr4_123282595_123282770 | 0.25 |
Pabpc4 |
poly(A) binding protein, cytoplasmic 4 |
143 |
0.91 |
| chr19_44418157_44418318 | 0.25 |
Gm50337 |
predicted gene, 50337 |
1373 |
0.35 |
| chr11_120316158_120316318 | 0.25 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
26445 |
0.07 |
| chr5_62622955_62623325 | 0.25 |
Gm43237 |
predicted gene 43237 |
9682 |
0.2 |
| chr4_32250961_32251192 | 0.25 |
Bach2it1 |
BTB and CNC homology 2, intronic transcript 1 |
685 |
0.69 |
| chrX_20635839_20636020 | 0.25 |
Rbm10 |
RNA binding motif protein 10 |
15831 |
0.11 |
| chr3_108297146_108297319 | 0.25 |
Gm43099 |
predicted gene 43099 |
9351 |
0.09 |
| chr1_133877032_133877215 | 0.24 |
Optc |
opticin |
24204 |
0.12 |
| chr4_46304170_46304340 | 0.24 |
Gm12447 |
predicted gene 12447 |
23041 |
0.12 |
| chr5_120514357_120514577 | 0.24 |
Slc8b1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
1324 |
0.29 |
| chr17_25863718_25863900 | 0.24 |
Mcrip2 |
MAPK regulated corepressor interacting protein 2 |
459 |
0.51 |
| chr7_105737131_105737562 | 0.24 |
Rrp8 |
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
39 |
0.72 |
| chr6_72142695_72142867 | 0.24 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
19584 |
0.11 |
| chr9_80688608_80688774 | 0.24 |
Gm39380 |
predicted gene, 39380 |
21304 |
0.24 |
| chr13_17805107_17805599 | 0.24 |
Cdk13 |
cyclin-dependent kinase 13 |
256 |
0.88 |
| chr16_18776692_18776858 | 0.24 |
Cldn5 |
claudin 5 |
72 |
0.96 |
| chr8_47713134_47713303 | 0.24 |
E030037K01Rik |
RIKEN cDNA E030037K01 gene |
48 |
0.91 |
| chr5_110431894_110432052 | 0.23 |
Fbrsl1 |
fibrosin-like 1 |
1348 |
0.35 |
| chr11_117778128_117778307 | 0.23 |
Tmc6 |
transmembrane channel-like gene family 6 |
1431 |
0.22 |
| chr12_111450305_111450614 | 0.23 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
60 |
0.96 |
| chr10_30000969_30001164 | 0.23 |
Gm48215 |
predicted gene, 48215 |
366 |
0.89 |
| chr9_44718034_44718200 | 0.23 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
1737 |
0.17 |
| chr19_7294990_7295178 | 0.23 |
Mark2 |
MAP/microtubule affinity regulating kinase 2 |
363 |
0.77 |
| chr9_110316019_110316191 | 0.23 |
Elp6 |
elongator acetyltransferase complex subunit 6 |
6458 |
0.11 |
| chr8_121950086_121950249 | 0.23 |
Banp |
BTG3 associated nuclear protein |
325 |
0.56 |
| chr4_106440974_106441125 | 0.23 |
Usp24 |
ubiquitin specific peptidase 24 |
12556 |
0.15 |
| chr17_45567292_45567749 | 0.22 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
2216 |
0.14 |
| chr13_54462200_54462359 | 0.22 |
Thoc3 |
THO complex 3 |
6509 |
0.16 |
| chr9_61319859_61320021 | 0.22 |
B930092H01Rik |
RIKEN cDNA B930092H01 gene |
26131 |
0.17 |
| chr1_75191952_75192286 | 0.22 |
Ankzf1 |
ankyrin repeat and zinc finger domain containing 1 |
32 |
0.52 |
| chr4_135241723_135242067 | 0.22 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
30919 |
0.12 |
| chr8_68083083_68083240 | 0.22 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
20875 |
0.24 |
| chr11_107342418_107342585 | 0.22 |
Gm11716 |
predicted gene 11716 |
4451 |
0.16 |
| chr11_70539975_70540306 | 0.22 |
Pld2 |
phospholipase D2 |
76 |
0.82 |
| chrX_129749622_129749793 | 0.22 |
Diaph2 |
diaphanous related formin 2 |
35 |
0.99 |
| chr7_101393766_101393917 | 0.22 |
Arap1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
527 |
0.67 |
| chr1_60662412_60662596 | 0.22 |
Gm11577 |
predicted gene 11577 |
38368 |
0.1 |
| chr5_3466112_3466325 | 0.21 |
Cdk6 |
cyclin-dependent kinase 6 |
4731 |
0.16 |
| chr9_103365880_103366048 | 0.21 |
Cdv3 |
carnitine deficiency-associated gene expressed in ventricle 3 |
124 |
0.95 |
| chr17_30611479_30611666 | 0.21 |
Glo1 |
glyoxalase 1 |
997 |
0.37 |
| chr11_16739244_16739507 | 0.21 |
Gm25698 |
predicted gene, 25698 |
6664 |
0.2 |
| chr7_80642020_80642180 | 0.21 |
Gm15880 |
predicted gene 15880 |
6083 |
0.17 |
| chr7_12997356_12997531 | 0.21 |
Slc27a5 |
solute carrier family 27 (fatty acid transporter), member 5 |
234 |
0.8 |
| chr19_4441877_4442205 | 0.21 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
1697 |
0.23 |
| chr7_141193568_141193999 | 0.21 |
Hras |
Harvey rat sarcoma virus oncogene |
99 |
0.77 |
| chr6_88761318_88761481 | 0.21 |
Gm43999 |
predicted gene, 43999 |
18672 |
0.11 |
| chr7_64357520_64357685 | 0.21 |
Gm20457 |
predicted gene 20457 |
11638 |
0.13 |
| chr11_114864022_114864192 | 0.21 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
385 |
0.8 |
| chr14_25181416_25181581 | 0.21 |
4930572O13Rik |
RIKEN cDNA 4930572O13 gene |
38257 |
0.14 |
| chr4_113008669_113008831 | 0.21 |
Gm12818 |
predicted gene 12818 |
28937 |
0.2 |
| chr1_164458750_164458928 | 0.21 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
484 |
0.74 |
| chr7_28982367_28982605 | 0.21 |
Map4k1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
342 |
0.62 |
| chr17_15192816_15193078 | 0.20 |
Gm35455 |
predicted gene, 35455 |
40788 |
0.13 |
| chr4_53595620_53595787 | 0.20 |
Slc44a1 |
solute carrier family 44, member 1 |
402 |
0.85 |
| chr17_12769492_12769649 | 0.20 |
Igf2r |
insulin-like growth factor 2 receptor |
94 |
0.95 |
| chr13_56645697_56645857 | 0.20 |
Tgfbi |
transforming growth factor, beta induced |
14564 |
0.19 |
| chr4_140621608_140621759 | 0.20 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
4618 |
0.21 |
| chr3_108287816_108288159 | 0.20 |
Gm43099 |
predicted gene 43099 |
106 |
0.92 |
| chr14_74732078_74732418 | 0.20 |
Esd |
esterase D/formylglutathione hydrolase |
49 |
0.98 |
| chr9_53323560_53323724 | 0.20 |
Exph5 |
exophilin 5 |
17442 |
0.18 |
| chr9_15058478_15058644 | 0.19 |
Panx1 |
pannexin 1 |
13083 |
0.18 |
| chr8_83580805_83581141 | 0.19 |
Gm45823 |
predicted gene 45823 |
4552 |
0.1 |
| chr2_58551945_58552231 | 0.19 |
Acvr1 |
activin A receptor, type 1 |
13650 |
0.2 |
| chr11_104463246_104463405 | 0.19 |
Kansl1 |
KAT8 regulatory NSL complex subunit 1 |
4666 |
0.18 |
| chr7_25706856_25707106 | 0.19 |
Ccdc97 |
coiled-coil domain containing 97 |
9125 |
0.09 |
| chr7_28867783_28867955 | 0.19 |
Lgals7 |
lectin, galactose binding, soluble 7 |
3414 |
0.11 |
| chr12_86883907_86884153 | 0.19 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
768 |
0.64 |
| chr13_41486979_41487189 | 0.19 |
Gm32401 |
predicted gene, 32401 |
46 |
0.65 |
| chr10_79865745_79866064 | 0.19 |
Plppr3 |
phospholipid phosphatase related 3 |
1377 |
0.14 |
| chr19_8251883_8252162 | 0.19 |
Gm41804 |
predicted gene, 41804 |
54 |
0.97 |
| chr15_78468233_78468783 | 0.19 |
Tmprss6 |
transmembrane serine protease 6 |
93 |
0.94 |
| chr8_106135686_106136171 | 0.19 |
1810019D21Rik |
RIKEN cDNA 1810019D21 gene |
493 |
0.53 |
| chr2_160840069_160840298 | 0.19 |
Gm11447 |
predicted gene 11447 |
5430 |
0.16 |
| chr3_104279647_104279910 | 0.18 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
59404 |
0.07 |
| chr8_115649918_115650129 | 0.18 |
Gm15655 |
predicted gene 15655 |
55055 |
0.12 |
| chr3_61363955_61364286 | 0.18 |
B430305J03Rik |
RIKEN cDNA B430305J03 gene |
1831 |
0.25 |
| chr1_153501584_153501799 | 0.18 |
Dhx9 |
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
14031 |
0.16 |
| chr9_14275987_14276328 | 0.18 |
Sesn3 |
sestrin 3 |
144 |
0.95 |
| chr6_3182898_3183288 | 0.18 |
Gm42642 |
predicted gene 42642 |
109 |
0.95 |
| chr11_98774757_98774908 | 0.18 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
501 |
0.64 |
| chr14_20929184_20929360 | 0.18 |
Vcl |
vinculin |
126 |
0.97 |
| chr16_10347675_10347844 | 0.18 |
Gm1600 |
predicted gene 1600 |
168 |
0.94 |
| chr8_90908060_90908476 | 0.17 |
Gm6658 |
predicted gene 6658 |
147 |
0.73 |
| chr1_191450665_191450840 | 0.17 |
Gm32200 |
predicted gene, 32200 |
14296 |
0.14 |
| chr19_4232043_4232236 | 0.17 |
Pold4 |
polymerase (DNA-directed), delta 4 |
153 |
0.85 |
| chr11_16815536_16815687 | 0.17 |
Egfros |
epidermal growth factor receptor, opposite strand |
15091 |
0.21 |
| chr4_149416520_149416730 | 0.17 |
Ube4b |
ubiquitination factor E4B |
9280 |
0.13 |
| chr5_137501838_137501997 | 0.17 |
Pop7 |
processing of precursor 7, ribonuclease P family, (S. cerevisiae) |
508 |
0.45 |
| chr14_52278932_52279090 | 0.17 |
Tox4 |
TOX high mobility group box family member 4 |
135 |
0.61 |
| chr10_79689120_79689284 | 0.17 |
Gzmm |
granzyme M (lymphocyte met-ase 1) |
182 |
0.86 |
| chr19_6349216_6349557 | 0.17 |
Map4k2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
1523 |
0.17 |
| chr10_128981137_128981300 | 0.17 |
Olfr9 |
olfactory receptor 9 |
8213 |
0.09 |
| chr8_46491538_46491689 | 0.17 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
639 |
0.67 |
| chr17_73995980_73996146 | 0.16 |
Gm24475 |
predicted gene, 24475 |
4797 |
0.19 |
| chr9_54843787_54843973 | 0.16 |
Rab7-ps1 |
RAB7, member RAS oncogene family, pseudogene 1 |
18598 |
0.15 |
| chr11_50354656_50355246 | 0.16 |
Cby3 |
chibby family member 3 |
489 |
0.68 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.3 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.2 | GO:2000412 | thymocyte migration(GO:0072679) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:1903911 | positive regulation of receptor clustering(GO:1903911) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.2 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:0045714 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.0 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.0 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.0 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.0 | GO:0009158 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.0 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0043814 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |