Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
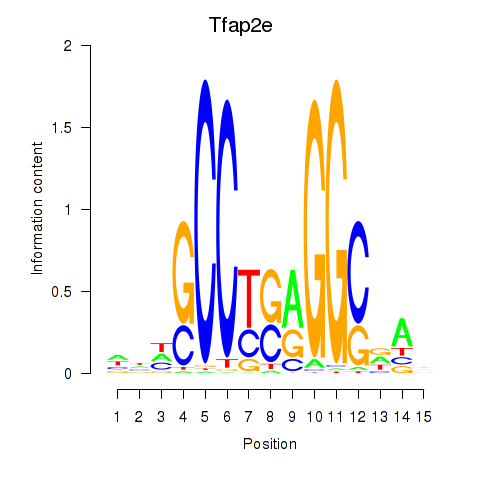
Results for Tfap2e
Z-value: 0.71
Transcription factors associated with Tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2e
|
ENSMUSG00000042477.7 | transcription factor AP-2, epsilon |
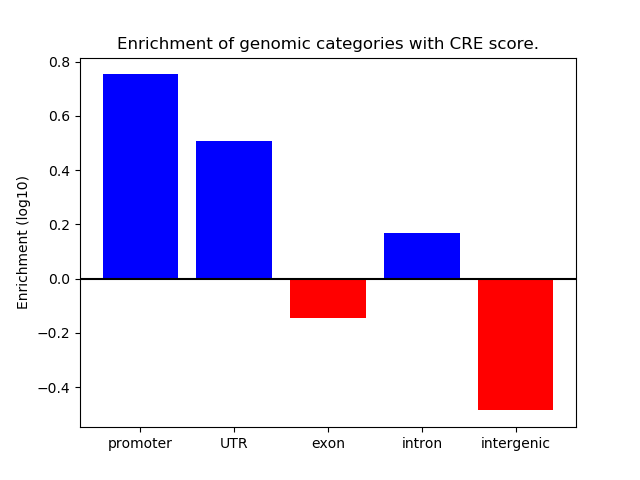
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_126733083_126733252 | Tfap2e | 3073 | 0.181558 | 0.71 | 1.1e-01 | Click! |
| chr4_126708617_126708772 | Tfap2e | 27546 | 0.107511 | 0.69 | 1.3e-01 | Click! |
| chr4_126741521_126741686 | Tfap2e | 5363 | 0.143797 | 0.07 | 8.9e-01 | Click! |
Activity of the Tfap2e motif across conditions
Conditions sorted by the z-value of the Tfap2e motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
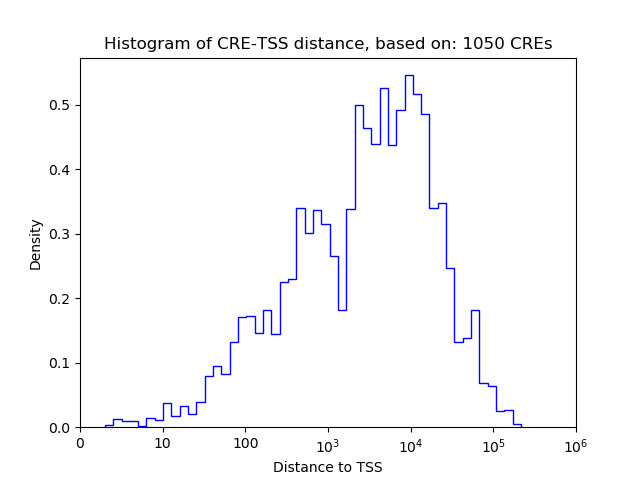
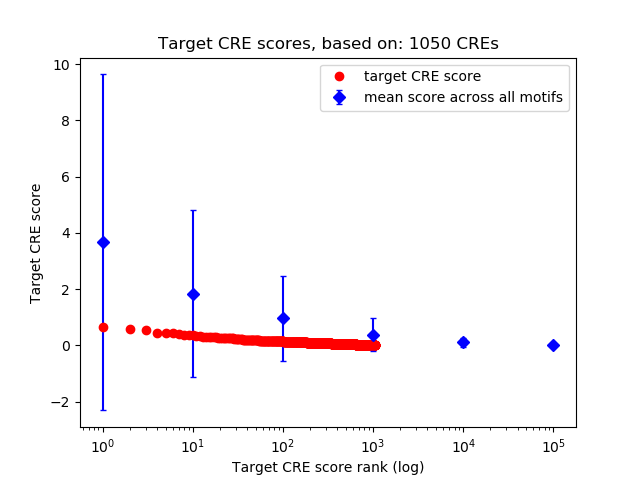
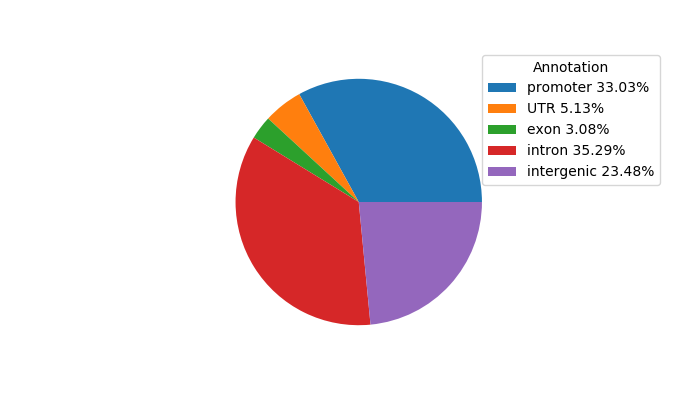
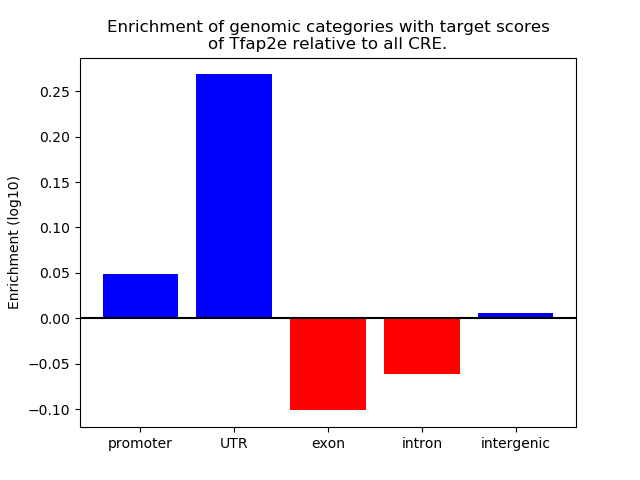
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_116636671_116636829 | 0.65 |
Rassf4 |
Ras association (RalGDS/AF-6) domain family member 4 |
4252 |
0.12 |
| chr12_112109668_112109994 | 0.58 |
Mir3073b |
microRNA 3073b |
522 |
0.45 |
| chr2_29750814_29750965 | 0.53 |
Gm13420 |
predicted gene 13420 |
796 |
0.47 |
| chr13_54402403_54402557 | 0.43 |
Gm16248 |
predicted gene 16248 |
3146 |
0.22 |
| chr9_115456234_115456417 | 0.43 |
Gm5921 |
predicted gene 5921 |
17744 |
0.16 |
| chr11_94390636_94390799 | 0.43 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
2203 |
0.26 |
| chr17_56681920_56682093 | 0.39 |
Ranbp3 |
RAN binding protein 3 |
8622 |
0.11 |
| chr2_145813125_145813276 | 0.38 |
Rin2 |
Ras and Rab interactor 2 |
8950 |
0.22 |
| chr19_54071291_54071462 | 0.38 |
Gm50186 |
predicted gene, 50186 |
13183 |
0.18 |
| chr3_108782866_108783017 | 0.37 |
Stxbp3 |
syntaxin binding protein 3 |
14843 |
0.14 |
| chr5_117285355_117285646 | 0.33 |
Pebp1 |
phosphatidylethanolamine binding protein 1 |
2025 |
0.19 |
| chr12_102295217_102295408 | 0.33 |
Rin3 |
Ras and Rab interactor 3 |
11671 |
0.22 |
| chr13_32897014_32897418 | 0.30 |
Serpinb1c |
serine (or cysteine) peptidase inhibitor, clade B, member 1c |
941 |
0.49 |
| chr1_72833711_72833862 | 0.30 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
8464 |
0.22 |
| chr19_25387114_25387309 | 0.30 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
8092 |
0.22 |
| chr17_29045019_29045203 | 0.29 |
Srsf3 |
serine and arginine-rich splicing factor 3 |
5417 |
0.09 |
| chr10_80957966_80958144 | 0.28 |
Gm3828 |
predicted gene 3828 |
3345 |
0.13 |
| chr9_95580937_95581120 | 0.28 |
Paqr9 |
progestin and adipoQ receptor family member IX |
21371 |
0.11 |
| chr11_64599304_64599471 | 0.27 |
Gm24275 |
predicted gene, 24275 |
8747 |
0.3 |
| chr12_110992829_110992984 | 0.26 |
6030440G07Rik |
RIKEN cDNA 6030440G07 gene |
1492 |
0.27 |
| chr19_34772591_34773080 | 0.26 |
Gm50173 |
predicted gene, 50173 |
1046 |
0.4 |
| chr19_30331979_30332136 | 0.26 |
Gm24108 |
predicted gene, 24108 |
70540 |
0.11 |
| chr2_39003650_39003831 | 0.26 |
Rpl35 |
ribosomal protein L35 |
1303 |
0.23 |
| chr6_73221980_73222151 | 0.26 |
Dnah6 |
dynein, axonemal, heavy chain 6 |
414 |
0.72 |
| chr11_53419604_53419795 | 0.26 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
3471 |
0.1 |
| chr12_102437143_102437359 | 0.25 |
Lgmn |
legumain |
2562 |
0.25 |
| chr16_18877584_18877767 | 0.25 |
Hira |
histone cell cycle regulator |
263 |
0.79 |
| chr14_61037704_61037855 | 0.24 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
112 |
0.6 |
| chr2_153657078_153657257 | 0.24 |
Dnmt3bos |
DNA methyltransferase 3B, opposite strand |
6881 |
0.16 |
| chr16_23989631_23990067 | 0.23 |
Bcl6 |
B cell leukemia/lymphoma 6 |
997 |
0.42 |
| chr18_46716349_46716534 | 0.23 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
11588 |
0.13 |
| chr9_67614466_67614621 | 0.22 |
Tln2 |
talin 2 |
54840 |
0.12 |
| chr5_114889052_114889221 | 0.22 |
Oasl2 |
2'-5' oligoadenylate synthetase-like 2 |
7800 |
0.08 |
| chr2_129489606_129489767 | 0.22 |
Gm14040 |
predicted gene 14040 |
26121 |
0.15 |
| chr4_59548512_59548664 | 0.21 |
Ptbp3 |
polypyrimidine tract binding protein 3 |
149 |
0.84 |
| chr7_84167720_84167916 | 0.20 |
Gm22177 |
predicted gene, 22177 |
2747 |
0.21 |
| chr1_39591675_39591839 | 0.20 |
Gm5100 |
predicted gene 5100 |
6248 |
0.13 |
| chr2_154609281_154609723 | 0.20 |
Zfp341 |
zinc finger protein 341 |
3795 |
0.12 |
| chr12_84206331_84206483 | 0.20 |
Gm47447 |
predicted gene, 47447 |
9730 |
0.1 |
| chr11_61564668_61564834 | 0.20 |
Epn2 |
epsin 2 |
14907 |
0.14 |
| chr5_9043706_9043857 | 0.20 |
Gm40264 |
predicted gene, 40264 |
8657 |
0.15 |
| chr16_95827519_95827681 | 0.20 |
1600002D24Rik |
RIKEN cDNA 1600002D24 gene |
18170 |
0.16 |
| chr13_92469480_92469792 | 0.19 |
Fam151b |
family with sequence similarity 151, member B |
14377 |
0.17 |
| chr1_165768492_165768643 | 0.19 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
909 |
0.37 |
| chr17_63493519_63493696 | 0.19 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
854 |
0.7 |
| chr9_70657719_70657882 | 0.19 |
Mindy2 |
MINDY lysine 48 deubiquitinase 2 |
626 |
0.63 |
| chr4_31426757_31426908 | 0.19 |
Gm11922 |
predicted gene 11922 |
46360 |
0.2 |
| chr8_71680518_71680684 | 0.19 |
Jak3 |
Janus kinase 3 |
2266 |
0.14 |
| chr3_145780988_145781495 | 0.18 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
20493 |
0.19 |
| chr19_41911790_41911957 | 0.18 |
Pgam1 |
phosphoglycerate mutase 1 |
50 |
0.95 |
| chr5_138779730_138779904 | 0.18 |
Fam20c |
family with sequence similarity 20, member C |
6683 |
0.2 |
| chr10_94119877_94120030 | 0.18 |
Gm48372 |
predicted gene, 48372 |
11214 |
0.13 |
| chr1_190174350_190174513 | 0.17 |
Prox1 |
prospero homeobox 1 |
3717 |
0.22 |
| chr19_34804924_34805353 | 0.17 |
Gm50116 |
predicted gene, 50116 |
876 |
0.48 |
| chr19_12634142_12634556 | 0.17 |
Glyat |
glycine-N-acyltransferase |
816 |
0.44 |
| chr19_6952737_6952905 | 0.17 |
Plcb3 |
phospholipase C, beta 3 |
2026 |
0.12 |
| chr14_73382885_73383301 | 0.17 |
Itm2b |
integral membrane protein 2B |
2105 |
0.33 |
| chr13_60493839_60494263 | 0.17 |
A530001N23Rik |
RIKEN cDNA A530001N23 gene |
5243 |
0.18 |
| chr5_88929617_88929827 | 0.17 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
4810 |
0.28 |
| chr18_46698271_46698422 | 0.17 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
29683 |
0.11 |
| chr5_32092059_32092277 | 0.17 |
Babam2 |
BRISC and BRCA1 A complex member 2 |
12471 |
0.17 |
| chr9_33068372_33068541 | 0.16 |
Gm27166 |
predicted gene 27166 |
36665 |
0.17 |
| chr19_4821405_4821565 | 0.16 |
Ccs |
copper chaperone for superoxide dismutase |
4524 |
0.09 |
| chr3_95625229_95625406 | 0.16 |
Ensa |
endosulfine alpha |
291 |
0.8 |
| chr18_31884190_31884397 | 0.16 |
Wdr33 |
WD repeat domain 33 |
2151 |
0.25 |
| chr10_84811233_84811541 | 0.16 |
Gm24226 |
predicted gene, 24226 |
204 |
0.95 |
| chr2_148872952_148873130 | 0.16 |
Cst3 |
cystatin C |
2410 |
0.19 |
| chr5_122274917_122275081 | 0.16 |
Pptc7 |
PTC7 protein phosphatase homolog |
9366 |
0.11 |
| chr18_12647726_12648094 | 0.15 |
Gm41668 |
predicted gene, 41668 |
509 |
0.72 |
| chr3_83994327_83994478 | 0.15 |
Tmem131l |
transmembrane 131 like |
26174 |
0.21 |
| chr12_71719042_71719430 | 0.15 |
Gm47555 |
predicted gene, 47555 |
35149 |
0.17 |
| chr2_119355713_119355881 | 0.15 |
Chac1 |
ChaC, cation transport regulator 1 |
4568 |
0.16 |
| chr9_51961247_51961723 | 0.15 |
Fdx1 |
ferredoxin 1 |
2061 |
0.29 |
| chr10_7212691_7212859 | 0.15 |
Cnksr3 |
Cnksr family member 3 |
538 |
0.86 |
| chr10_77693450_77693640 | 0.15 |
Gm19402 |
predicted gene, 19402 |
2788 |
0.08 |
| chr5_99994276_99994439 | 0.15 |
Gm17092 |
predicted gene 17092 |
15296 |
0.13 |
| chr12_78265748_78266376 | 0.15 |
Gm48225 |
predicted gene, 48225 |
4769 |
0.18 |
| chr10_75222636_75222997 | 0.15 |
Specc1l |
sperm antigen with calponin homology and coiled-coil domains 1-like |
7039 |
0.21 |
| chr13_46883271_46883538 | 0.15 |
Gm48250 |
predicted gene, 48250 |
2377 |
0.23 |
| chr9_43467006_43467175 | 0.15 |
Gm28215 |
predicted gene 28215 |
2332 |
0.29 |
| chr6_117612056_117612213 | 0.15 |
Gm45083 |
predicted gene 45083 |
436 |
0.85 |
| chr14_20298249_20298415 | 0.14 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
2170 |
0.22 |
| chr17_25221206_25221426 | 0.14 |
Unkl |
unkempt family like zinc finger |
1223 |
0.25 |
| chr4_57845057_57845232 | 0.14 |
Pakap |
paralemmin A kinase anchor protein |
103 |
0.97 |
| chr5_118596025_118596197 | 0.14 |
Gm43788 |
predicted gene 43788 |
6605 |
0.16 |
| chr2_104002844_104003003 | 0.14 |
Fbxo3 |
F-box protein 3 |
24798 |
0.1 |
| chr4_135989135_135989286 | 0.14 |
Pithd1 |
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
1946 |
0.16 |
| chr8_11636365_11636526 | 0.14 |
Ankrd10 |
ankyrin repeat domain 10 |
688 |
0.29 |
| chr7_139364090_139364263 | 0.14 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
24933 |
0.19 |
| chr15_102721608_102721977 | 0.14 |
Calcoco1 |
calcium binding and coiled coil domain 1 |
339 |
0.84 |
| chr11_7196610_7196788 | 0.14 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1083 |
0.48 |
| chr11_95687340_95687492 | 0.14 |
Gm9796 |
predicted gene 9796 |
11727 |
0.12 |
| chr18_32136413_32136564 | 0.14 |
Proc |
protein C |
742 |
0.51 |
| chr1_164307490_164307653 | 0.14 |
Blzf1 |
basic leucine zipper nuclear factor 1 |
82 |
0.52 |
| chr1_69106645_69106814 | 0.14 |
Gm16076 |
predicted gene 16076 |
11 |
0.96 |
| chr10_128298417_128298592 | 0.14 |
Il23a |
interleukin 23, alpha subunit p19 |
248 |
0.74 |
| chr7_27478150_27478347 | 0.14 |
Sertad3 |
SERTA domain containing 3 |
4480 |
0.1 |
| chr13_83064851_83065002 | 0.14 |
2310067P03Rik |
RIKEN cDNA 2310067P03 gene |
531 |
0.88 |
| chr6_73222805_73223119 | 0.14 |
Gm40377 |
predicted gene, 40377 |
88 |
0.95 |
| chr1_190847941_190848107 | 0.14 |
Rps6kc1 |
ribosomal protein S6 kinase polypeptide 1 |
3440 |
0.28 |
| chr11_121500753_121500917 | 0.14 |
Tbcd |
tubulin-specific chaperone d |
799 |
0.61 |
| chr2_160210649_160210800 | 0.13 |
Gm826 |
predicted gene 826 |
102892 |
0.07 |
| chr16_30255365_30255526 | 0.13 |
Gm49645 |
predicted gene, 49645 |
293 |
0.87 |
| chr17_32364458_32364626 | 0.13 |
Wiz |
widely-interspaced zinc finger motifs |
79 |
0.95 |
| chr19_6243342_6243519 | 0.13 |
Atg2a |
autophagy related 2A |
1762 |
0.16 |
| chr17_79715662_79715994 | 0.13 |
Cyp1b1 |
cytochrome P450, family 1, subfamily b, polypeptide 1 |
767 |
0.62 |
| chr14_31929383_31929549 | 0.13 |
D830044D21Rik |
RIKEN cDNA D830044D21 gene |
22938 |
0.17 |
| chr8_126685352_126685531 | 0.13 |
Gm45805 |
predicted gene 45805 |
72893 |
0.1 |
| chr9_80026313_80026485 | 0.13 |
Filip1 |
filamin A interacting protein 1 |
13548 |
0.14 |
| chr1_166032024_166032183 | 0.13 |
Gm7626 |
predicted gene 7626 |
11588 |
0.15 |
| chr9_64053601_64053804 | 0.13 |
Gm25606 |
predicted gene, 25606 |
5206 |
0.14 |
| chr10_115479053_115479225 | 0.13 |
Lgr5 |
leucine rich repeat containing G protein coupled receptor 5 |
16060 |
0.19 |
| chr15_85709424_85709582 | 0.13 |
Mirlet7b |
microRNA let7b |
2184 |
0.21 |
| chr9_46053003_46053558 | 0.13 |
Sik3 |
SIK family kinase 3 |
40280 |
0.11 |
| chr11_75689615_75689766 | 0.13 |
Crk |
v-crk avian sarcoma virus CT10 oncogene homolog |
10148 |
0.13 |
| chr4_44997094_44997250 | 0.13 |
1700055D18Rik |
RIKEN cDNA 1700055D18 gene |
14515 |
0.1 |
| chr4_145037079_145037530 | 0.13 |
Vps13d |
vacuolar protein sorting 13D |
13730 |
0.24 |
| chr4_139373277_139373435 | 0.13 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
7303 |
0.1 |
| chr6_88292396_88292777 | 0.13 |
Gm44265 |
predicted gene, 44265 |
61020 |
0.08 |
| chr4_104973013_104973233 | 0.13 |
Gm12721 |
predicted gene 12721 |
2994 |
0.33 |
| chr8_71375544_71375702 | 0.13 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
167 |
0.89 |
| chr10_69288238_69288393 | 0.13 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
8857 |
0.18 |
| chr4_136248818_136248972 | 0.12 |
Tcea3 |
transcription elongation factor A (SII), 3 |
922 |
0.5 |
| chr6_108254587_108254738 | 0.12 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
1704 |
0.47 |
| chrX_100447303_100447475 | 0.12 |
Awat2 |
acyl-CoA wax alcohol acyltransferase 2 |
4672 |
0.17 |
| chr1_72226394_72226550 | 0.12 |
Gm25360 |
predicted gene, 25360 |
232 |
0.89 |
| chr9_62421178_62421341 | 0.12 |
Coro2b |
coronin, actin binding protein, 2B |
6742 |
0.24 |
| chr11_86921243_86921467 | 0.12 |
Ypel2 |
yippee like 2 |
50669 |
0.12 |
| chr8_70221071_70221246 | 0.12 |
Armc6 |
armadillo repeat containing 6 |
1132 |
0.32 |
| chr18_38180760_38180944 | 0.12 |
Pcdh1 |
protocadherin 1 |
22311 |
0.12 |
| chr8_124750214_124750687 | 0.12 |
Fam89a |
family with sequence similarity 89, member A |
1382 |
0.3 |
| chr8_93043146_93043627 | 0.12 |
Ces1a |
carboxylesterase 1A |
2283 |
0.25 |
| chr7_25390104_25390268 | 0.12 |
Lipe |
lipase, hormone sensitive |
80 |
0.94 |
| chr4_141131590_141132080 | 0.12 |
Szrd1 |
SUZ RNA binding domain containing 1 |
7892 |
0.11 |
| chr1_161247888_161248058 | 0.12 |
Prdx6 |
peroxiredoxin 6 |
2581 |
0.25 |
| chr10_77118262_77118451 | 0.12 |
Col18a1 |
collagen, type XVIII, alpha 1 |
4410 |
0.2 |
| chr18_20188219_20188403 | 0.12 |
Dsg1c |
desmoglein 1 gamma |
59029 |
0.12 |
| chr5_139499620_139499800 | 0.12 |
Zfand2a |
zinc finger, AN1-type domain 2A |
15161 |
0.13 |
| chr9_40805180_40805541 | 0.12 |
Snord14e |
small nucleolar RNA, C/D box 14E |
612 |
0.38 |
| chr12_30943371_30943530 | 0.12 |
Sh3yl1 |
Sh3 domain YSC-like 1 |
31263 |
0.14 |
| chr5_34187720_34187884 | 0.12 |
Mxd4 |
Max dimerization protein 4 |
82 |
0.94 |
| chr14_29996362_29996748 | 0.12 |
Il17rb |
interleukin 17 receptor B |
1904 |
0.2 |
| chr8_84743526_84743857 | 0.12 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
20684 |
0.09 |
| chr12_104841328_104841498 | 0.11 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
15958 |
0.18 |
| chr10_41575979_41576291 | 0.11 |
5730435O14Rik |
RIKEN cDNA 5730435O14 gene |
571 |
0.67 |
| chr11_119913012_119913281 | 0.11 |
Chmp6 |
charged multivesicular body protein 6 |
295 |
0.86 |
| chr10_67025749_67025925 | 0.11 |
Gm31763 |
predicted gene, 31763 |
2528 |
0.28 |
| chr3_104360355_104360770 | 0.11 |
Gm5546 |
predicted gene 5546 |
6051 |
0.18 |
| chr17_25239677_25239863 | 0.11 |
Gnptg |
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
346 |
0.47 |
| chr11_50137654_50137855 | 0.11 |
Tbc1d9b |
TBC1 domain family, member 9B |
2327 |
0.18 |
| chr8_22124586_22124925 | 0.11 |
Nek5 |
NIMA (never in mitosis gene a)-related expressed kinase 5 |
283 |
0.87 |
| chr8_84346540_84346704 | 0.11 |
Cacna1a |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
7983 |
0.14 |
| chr15_100637292_100637581 | 0.11 |
Smagp |
small cell adhesion glycoprotein |
496 |
0.61 |
| chr8_70775755_70775922 | 0.11 |
Pik3r2 |
phosphoinositide-3-kinase regulatory subunit 2 |
841 |
0.26 |
| chr10_84416787_84416966 | 0.11 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
23721 |
0.15 |
| chr5_122263936_122264091 | 0.11 |
Tctn1 |
tectonic family member 1 |
402 |
0.76 |
| chr12_46635586_46635767 | 0.11 |
Gm48535 |
predicted gene, 48535 |
30033 |
0.15 |
| chr9_80777245_80777426 | 0.11 |
Gm27224 |
predicted gene 27224 |
25316 |
0.22 |
| chr8_126808574_126808734 | 0.11 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
30579 |
0.17 |
| chr18_69502311_69502466 | 0.11 |
Tcf4 |
transcription factor 4 |
1430 |
0.53 |
| chr19_46975598_46975758 | 0.11 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
6110 |
0.17 |
| chr13_75598831_75599007 | 0.11 |
Gm4149 |
predicted pseudogene 4149 |
45977 |
0.1 |
| chr4_62508801_62509208 | 0.11 |
Hdhd3 |
haloacid dehalogenase-like hydrolase domain containing 3 |
5537 |
0.12 |
| chr18_38283827_38284012 | 0.11 |
Pcdh12 |
protocadherin 12 |
438 |
0.52 |
| chr12_40580026_40580177 | 0.11 |
Dock4 |
dedicator of cytokinesis 4 |
133765 |
0.05 |
| chr1_58112957_58113355 | 0.11 |
Aox3 |
aldehyde oxidase 3 |
18 |
0.98 |
| chr13_67780014_67780192 | 0.11 |
Zfp493 |
zinc finger protein 493 |
349 |
0.76 |
| chr3_121291310_121291524 | 0.11 |
Alg14 |
asparagine-linked glycosylation 14 |
356 |
0.84 |
| chr6_99490333_99490484 | 0.11 |
Gm22328 |
predicted gene, 22328 |
5706 |
0.22 |
| chr17_14173768_14173919 | 0.10 |
Gm34510 |
predicted gene, 34510 |
29885 |
0.13 |
| chr14_66654420_66654619 | 0.10 |
Adra1a |
adrenergic receptor, alpha 1a |
18963 |
0.22 |
| chr4_135286870_135287040 | 0.10 |
Gm12983 |
predicted gene 12983 |
3720 |
0.15 |
| chr5_103738632_103738798 | 0.10 |
Aff1 |
AF4/FMR2 family, member 1 |
15447 |
0.19 |
| chr12_102647960_102648119 | 0.10 |
Itpk1 |
inositol 1,3,4-triphosphate 5/6 kinase |
7105 |
0.14 |
| chr15_77924420_77924584 | 0.10 |
Txn2 |
thioredoxin 2 |
3233 |
0.19 |
| chr9_74921528_74921682 | 0.10 |
Fam214a |
family with sequence similarity 214, member A |
31279 |
0.14 |
| chr17_31391456_31391613 | 0.10 |
Pde9a |
phosphodiesterase 9A |
5240 |
0.16 |
| chr5_114581660_114581827 | 0.10 |
Fam222a |
family with sequence similarity 222, member A |
13726 |
0.15 |
| chr14_20138027_20138204 | 0.10 |
Saysd1 |
SAYSVFN motif domain containing 1 |
419 |
0.81 |
| chr10_24097646_24097797 | 0.10 |
Taar8c |
trace amine-associated receptor 8C |
4191 |
0.1 |
| chr4_148214912_148215063 | 0.10 |
Disp3 |
dispatched RND transporter family member 3 |
45764 |
0.08 |
| chr3_119783480_119783653 | 0.10 |
Ptbp2 |
polypyrimidine tract binding protein 2 |
125 |
0.96 |
| chr5_114435733_114435884 | 0.10 |
Mmab |
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human) |
8184 |
0.14 |
| chr5_74316665_74316828 | 0.10 |
Gm15981 |
predicted gene 15981 |
288 |
0.89 |
| chr8_119445523_119446899 | 0.10 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
508 |
0.74 |
| chr7_115565335_115565528 | 0.10 |
Sox6 |
SRY (sex determining region Y)-box 6 |
32465 |
0.24 |
| chr2_166952238_166952420 | 0.10 |
Stau1 |
staufen double-stranded RNA binding protein 1 |
1098 |
0.41 |
| chr1_10212196_10212367 | 0.10 |
Arfgef1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) |
1932 |
0.35 |
| chrX_20819269_20819429 | 0.10 |
Gm24824 |
predicted gene, 24824 |
4465 |
0.13 |
| chr6_94357642_94357793 | 0.10 |
Gm19908 |
predicted gene, 19908 |
8183 |
0.21 |
| chr15_96699087_96699266 | 0.10 |
Slc38a2 |
solute carrier family 38, member 2 |
485 |
0.66 |
| chr4_97772221_97772394 | 0.10 |
Nfia |
nuclear factor I/A |
427 |
0.84 |
| chr15_64152976_64153142 | 0.10 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
29253 |
0.18 |
| chr8_124275783_124275948 | 0.10 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
19238 |
0.16 |
| chr8_120295587_120295753 | 0.10 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
67214 |
0.09 |
| chr12_24627702_24627853 | 0.10 |
Gm6969 |
predicted pseudogene 6969 |
11233 |
0.15 |
| chr7_100658408_100658565 | 0.10 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
30 |
0.96 |
| chr17_13699350_13699759 | 0.09 |
Gm16046 |
predicted gene 16046 |
16038 |
0.13 |
| chr17_56470625_56470795 | 0.09 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
3917 |
0.18 |
| chr11_46579639_46579790 | 0.09 |
BC053393 |
cDNA sequence BC053393 |
8178 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |