Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
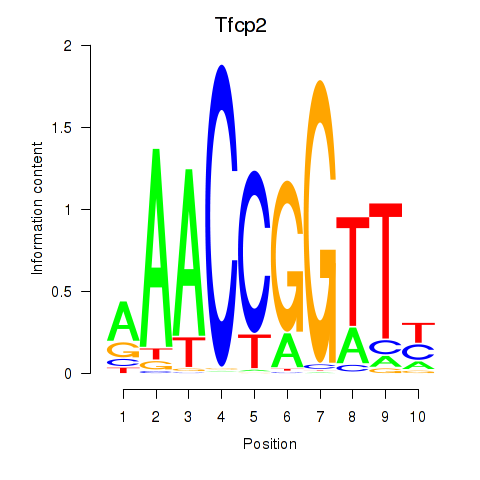
Results for Tfcp2
Z-value: 0.88
Transcription factors associated with Tfcp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2
|
ENSMUSG00000009733.8 | transcription factor CP2 |
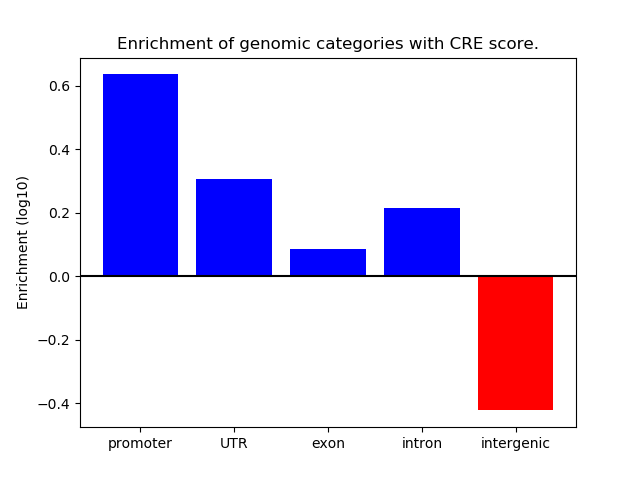
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_100537987_100538140 | Tfcp2 | 12330 | 0.108936 | -0.79 | 5.9e-02 | Click! |
| chr15_100551769_100552156 | Tfcp2 | 19 | 0.959085 | 0.54 | 2.7e-01 | Click! |
| chr15_100551472_100551651 | Tfcp2 | 10 | 0.959615 | -0.05 | 9.3e-01 | Click! |
| chr15_100552258_100552413 | Tfcp2 | 327 | 0.808787 | -0.03 | 9.5e-01 | Click! |
Activity of the Tfcp2 motif across conditions
Conditions sorted by the z-value of the Tfcp2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
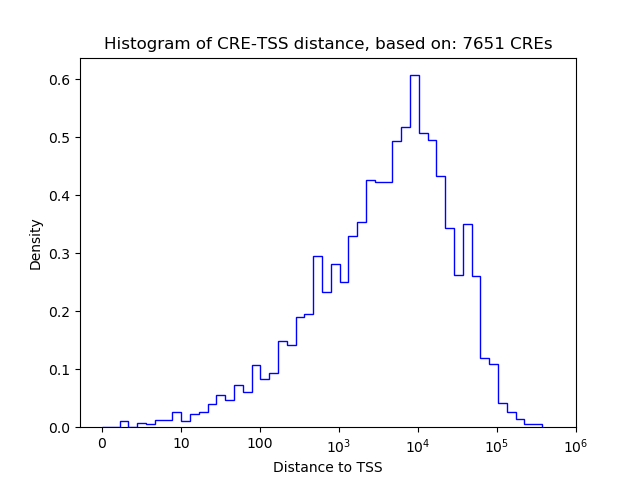
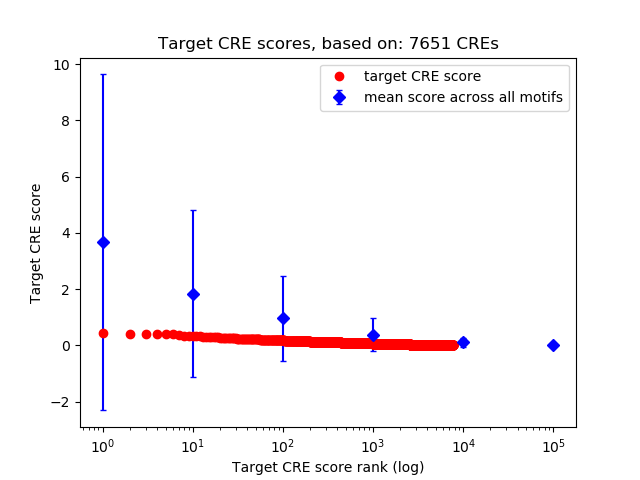
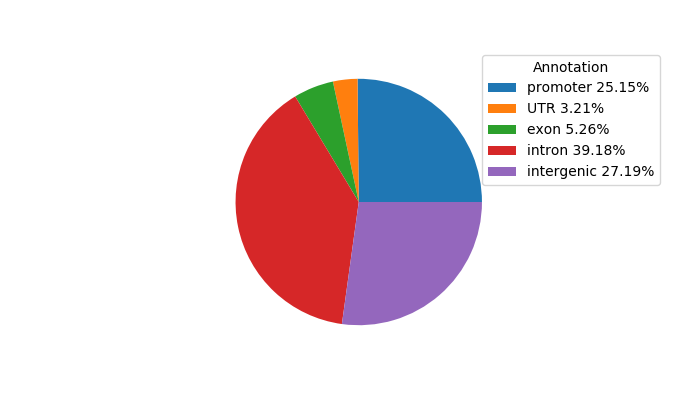
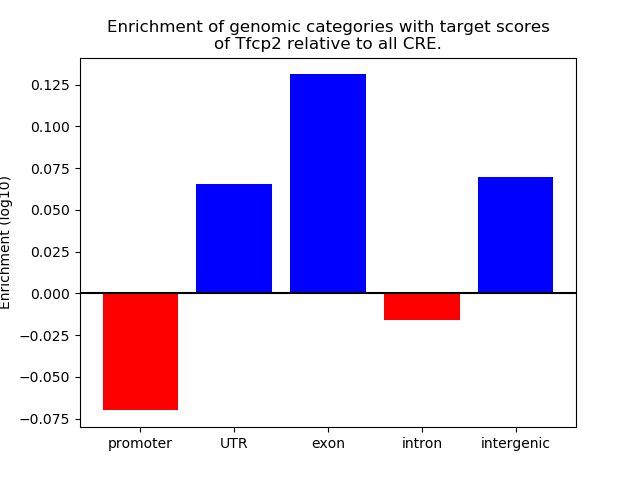
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_16530464_16530632 | 0.44 |
Rarb |
retinoic acid receptor, beta |
44497 |
0.15 |
| chr19_43971247_43971535 | 0.42 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr15_68255673_68255836 | 0.42 |
Zfat |
zinc finger and AT hook domain containing |
3033 |
0.26 |
| chr6_128504140_128504367 | 0.41 |
Pzp |
PZP, alpha-2-macroglobulin like |
9030 |
0.09 |
| chr6_51392183_51392564 | 0.40 |
0610033M10Rik |
RIKEN cDNA 0610033M10 gene |
9660 |
0.17 |
| chr2_76639734_76639916 | 0.39 |
Prkra |
protein kinase, interferon inducible double stranded RNA dependent activator |
908 |
0.53 |
| chr4_117342819_117343008 | 0.37 |
Rnf220 |
ring finger protein 220 |
32544 |
0.12 |
| chr5_120514188_120514356 | 0.35 |
Slc8b1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
1129 |
0.34 |
| chr17_27557519_27557704 | 0.35 |
Hmga1 |
high mobility group AT-hook 1 |
916 |
0.28 |
| chr10_95878338_95878489 | 0.34 |
Gm47724 |
predicted gene, 47724 |
7614 |
0.13 |
| chr6_54759739_54759890 | 0.33 |
Znrf2 |
zinc and ring finger 2 |
57102 |
0.1 |
| chr14_30890557_30890708 | 0.32 |
Itih4 |
inter alpha-trypsin inhibitor, heavy chain 4 |
4111 |
0.13 |
| chr5_8977445_8977723 | 0.31 |
Crot |
carnitine O-octanoyltransferase |
4560 |
0.14 |
| chr5_88918674_88918881 | 0.30 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
15755 |
0.23 |
| chr7_141171399_141171582 | 0.30 |
Rnh1 |
ribonuclease/angiogenin inhibitor 1 |
1367 |
0.2 |
| chr17_3989752_3989920 | 0.30 |
4930470H14Rik |
RIKEN cDNA 4930470H14 gene |
93159 |
0.09 |
| chr1_64791801_64792159 | 0.29 |
Plekhm3 |
pleckstrin homology domain containing, family M, member 3 |
44784 |
0.11 |
| chr4_60659551_60660018 | 0.29 |
Mup11 |
major urinary protein 11 |
46 |
0.97 |
| chr8_94172420_94173095 | 0.28 |
Mt2 |
metallothionein 2 |
93 |
0.88 |
| chr8_119425498_119425673 | 0.26 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
8539 |
0.15 |
| chr4_15327477_15327645 | 0.26 |
Gm24317 |
predicted gene, 24317 |
55587 |
0.13 |
| chr10_13869659_13869820 | 0.26 |
Aig1 |
androgen-induced 1 |
759 |
0.5 |
| chr5_145873496_145873886 | 0.26 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
3000 |
0.22 |
| chr12_21183631_21184095 | 0.26 |
AC156032.1 |
|
63460 |
0.08 |
| chr2_72881753_72881905 | 0.25 |
Sp3 |
trans-acting transcription factor 3 |
59255 |
0.1 |
| chr12_79140897_79141048 | 0.25 |
Arg2 |
arginase type II |
10195 |
0.09 |
| chr4_35134107_35134262 | 0.25 |
Ifnk |
interferon kappa |
17872 |
0.17 |
| chr6_91111481_91112156 | 0.25 |
Nup210 |
nucleoporin 210 |
4978 |
0.17 |
| chr1_165766009_165766175 | 0.25 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
2346 |
0.15 |
| chr7_27542323_27542474 | 0.24 |
Pld3 |
phospholipase D family, member 3 |
347 |
0.77 |
| chr4_57312348_57312552 | 0.24 |
Ptpn3 |
protein tyrosine phosphatase, non-receptor type 3 |
10613 |
0.17 |
| chr6_94357642_94357793 | 0.23 |
Gm19908 |
predicted gene, 19908 |
8183 |
0.21 |
| chr9_70913787_70914085 | 0.23 |
Gm32017 |
predicted gene, 32017 |
16552 |
0.19 |
| chr12_101766669_101766841 | 0.23 |
Tc2n |
tandem C2 domains, nuclear |
48232 |
0.12 |
| chr1_67194458_67194637 | 0.23 |
Gm15668 |
predicted gene 15668 |
54653 |
0.12 |
| chr11_101377034_101377689 | 0.23 |
G6pc |
glucose-6-phosphatase, catalytic |
9800 |
0.06 |
| chr9_108785280_108785665 | 0.23 |
Ip6k2 |
inositol hexaphosphate kinase 2 |
1607 |
0.21 |
| chr2_170287488_170287839 | 0.23 |
Gm14270 |
predicted gene 14270 |
2628 |
0.33 |
| chr10_63160777_63160928 | 0.23 |
Mypn |
myopalladin |
38168 |
0.1 |
| chr12_77216600_77216773 | 0.22 |
Fut8 |
fucosyltransferase 8 |
21439 |
0.19 |
| chr1_16172552_16172725 | 0.22 |
Gm38249 |
predicted gene, 38249 |
37879 |
0.13 |
| chr8_34434777_34434932 | 0.22 |
Gm45349 |
predicted gene 45349 |
38141 |
0.12 |
| chr12_7987781_7988288 | 0.22 |
Apob |
apolipoprotein B |
10255 |
0.22 |
| chr17_12415417_12415751 | 0.22 |
Plg |
plasminogen |
36925 |
0.12 |
| chr10_93491843_93492023 | 0.22 |
Hal |
histidine ammonia lyase |
3130 |
0.19 |
| chr1_105965333_105965503 | 0.22 |
Gm18801 |
predicted gene, 18801 |
9108 |
0.15 |
| chr1_36573916_36574068 | 0.22 |
Fam178b |
family with sequence similarity 178, member B |
13695 |
0.09 |
| chr4_60821413_60821822 | 0.21 |
Mup22 |
major urinary protein 22 |
154 |
0.96 |
| chr10_7976561_7976927 | 0.21 |
Tab2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
20514 |
0.19 |
| chr15_99406396_99406547 | 0.21 |
Tmbim6 |
transmembrane BAX inhibitor motif containing 6 |
597 |
0.56 |
| chr4_123576766_123577231 | 0.21 |
Macf1 |
microtubule-actin crosslinking factor 1 |
3938 |
0.22 |
| chr14_41104159_41104453 | 0.21 |
Mat1a |
methionine adenosyltransferase I, alpha |
729 |
0.55 |
| chr2_118534050_118534205 | 0.21 |
Bmf |
BCL2 modifying factor |
10460 |
0.16 |
| chr7_68717199_68717404 | 0.21 |
Gm44692 |
predicted gene 44692 |
9166 |
0.23 |
| chr15_86177362_86177531 | 0.21 |
Gm15569 |
predicted gene 15569 |
8282 |
0.17 |
| chr18_53353090_53353276 | 0.20 |
Ppic |
peptidylprolyl isomerase C |
64932 |
0.12 |
| chr5_90872752_90873224 | 0.20 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
18253 |
0.11 |
| chr8_94186443_94186638 | 0.20 |
Gm39228 |
predicted gene, 39228 |
3251 |
0.13 |
| chr13_37943133_37943379 | 0.20 |
Rreb1 |
ras responsive element binding protein 1 |
3760 |
0.25 |
| chr16_22914452_22914638 | 0.20 |
Fetub |
fetuin beta |
3789 |
0.14 |
| chr19_3958627_3958778 | 0.20 |
Aldh3b3 |
aldehyde dehydrogenase 3 family, member B3 |
57 |
0.92 |
| chr13_30233563_30233739 | 0.20 |
Mboat1 |
membrane bound O-acyltransferase domain containing 1 |
1798 |
0.39 |
| chr11_64844149_64844351 | 0.20 |
Gm12292 |
predicted gene 12292 |
497 |
0.87 |
| chr5_117245335_117245486 | 0.20 |
Taok3 |
TAO kinase 3 |
4950 |
0.16 |
| chr9_32718091_32718269 | 0.20 |
Gm27240 |
predicted gene 27240 |
7009 |
0.2 |
| chr1_191225741_191225922 | 0.20 |
D730003I15Rik |
RIKEN cDNA D730003I15 gene |
1357 |
0.33 |
| chr8_117718222_117718447 | 0.20 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
2602 |
0.2 |
| chr14_100324093_100324244 | 0.20 |
Gm41231 |
predicted gene, 41231 |
38003 |
0.15 |
| chr1_178330385_178330898 | 0.20 |
Hnrnpu |
heterogeneous nuclear ribonucleoprotein U |
20 |
0.96 |
| chr10_93742676_93742904 | 0.20 |
Gm15963 |
predicted gene 15963 |
14583 |
0.15 |
| chr7_44892625_44892974 | 0.20 |
Med25 |
mediator complex subunit 25 |
87 |
0.91 |
| chr14_73445592_73445761 | 0.19 |
Gm4266 |
predicted gene 4266 |
47417 |
0.11 |
| chr4_60741366_60741658 | 0.19 |
Mup12 |
major urinary protein 12 |
186 |
0.94 |
| chr9_58085168_58085353 | 0.19 |
Ccdc33 |
coiled-coil domain containing 33 |
3191 |
0.18 |
| chr9_78031227_78031411 | 0.19 |
Gm47829 |
predicted gene, 47829 |
3374 |
0.14 |
| chr16_31493024_31493411 | 0.19 |
Gm46560 |
predicted gene, 46560 |
15170 |
0.14 |
| chr10_24635350_24635521 | 0.19 |
Gm15270 |
predicted gene 15270 |
38327 |
0.13 |
| chr8_33913458_33913624 | 0.19 |
Rbpms |
RNA binding protein gene with multiple splicing |
15735 |
0.16 |
| chr4_145041587_145041938 | 0.19 |
Vps13d |
vacuolar protein sorting 13D |
9272 |
0.25 |
| chr7_123193753_123193930 | 0.19 |
Tnrc6a |
trinucleotide repeat containing 6a |
13920 |
0.2 |
| chr5_118246346_118246796 | 0.19 |
2410131K14Rik |
RIKEN cDNA 2410131K14 gene |
1344 |
0.32 |
| chr11_107130421_107130628 | 0.19 |
Bptf |
bromodomain PHD finger transcription factor |
1398 |
0.36 |
| chr11_115290095_115290276 | 0.19 |
Fads6 |
fatty acid desaturase domain family, member 6 |
7329 |
0.1 |
| chr6_142752473_142752975 | 0.19 |
Cmas |
cytidine monophospho-N-acetylneuraminic acid synthetase |
3962 |
0.23 |
| chr14_45457123_45457466 | 0.19 |
Gm34250 |
predicted gene, 34250 |
10221 |
0.12 |
| chr13_52236242_52236425 | 0.19 |
Gm48199 |
predicted gene, 48199 |
55922 |
0.15 |
| chr1_36527353_36527514 | 0.18 |
Gm38033 |
predicted gene, 38033 |
804 |
0.4 |
| chr11_121082992_121083151 | 0.18 |
Sectm1a |
secreted and transmembrane 1A |
1851 |
0.17 |
| chr1_184060517_184060686 | 0.18 |
Dusp10 |
dual specificity phosphatase 10 |
26220 |
0.19 |
| chr5_102483486_102483664 | 0.18 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
286 |
0.9 |
| chr3_98322281_98322464 | 0.18 |
Phgdh |
3-phosphoglycerate dehydrogenase |
6241 |
0.17 |
| chr11_69110217_69110963 | 0.18 |
Hes7 |
hes family bHLH transcription factor 7 |
9814 |
0.07 |
| chr15_89209095_89209277 | 0.18 |
Ppp6r2 |
protein phosphatase 6, regulatory subunit 2 |
2367 |
0.16 |
| chr3_95668091_95668297 | 0.18 |
Adamtsl4 |
ADAMTS-like 4 |
8957 |
0.1 |
| chr13_93648931_93649116 | 0.18 |
Bhmt |
betaine-homocysteine methyltransferase |
11062 |
0.14 |
| chr5_32141078_32141248 | 0.18 |
Fosl2 |
fos-like antigen 2 |
4990 |
0.17 |
| chr7_139247418_139247589 | 0.17 |
Pwwp2b |
PWWP domain containing 2B |
979 |
0.48 |
| chr14_120318430_120318607 | 0.17 |
Mbnl2 |
muscleblind like splicing factor 2 |
12062 |
0.26 |
| chr15_83704064_83704215 | 0.17 |
Scube1 |
signal peptide, CUB domain, EGF-like 1 |
20787 |
0.2 |
| chr12_31295518_31295688 | 0.17 |
Lamb1 |
laminin B1 |
17428 |
0.13 |
| chr10_77086952_77087277 | 0.17 |
Col18a1 |
collagen, type XVIII, alpha 1 |
2314 |
0.26 |
| chr5_8940920_8941071 | 0.17 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
12269 |
0.13 |
| chr11_94207284_94207435 | 0.17 |
Tob1 |
transducer of ErbB-2.1 |
4095 |
0.21 |
| chr2_156451427_156451737 | 0.17 |
Gm14225 |
predicted gene 14225 |
3974 |
0.13 |
| chr6_129434345_129434496 | 0.17 |
Gm43914 |
predicted gene, 43914 |
16241 |
0.09 |
| chr2_158225371_158225751 | 0.17 |
D630003M21Rik |
RIKEN cDNA D630003M21 gene |
3661 |
0.17 |
| chr17_83503490_83503653 | 0.17 |
Cox7a2l |
cytochrome c oxidase subunit 7A2 like |
935 |
0.66 |
| chr6_128604604_128605019 | 0.17 |
Gm44009 |
predicted gene, 44009 |
16786 |
0.08 |
| chr1_178230411_178230566 | 0.17 |
Desi2 |
desumoylating isopeptidase 2 |
25615 |
0.13 |
| chr5_149247178_149247337 | 0.17 |
Gm29264 |
predicted gene 29264 |
425 |
0.57 |
| chr1_132365008_132365179 | 0.17 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
145 |
0.93 |
| chr9_57287321_57287487 | 0.17 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
24792 |
0.13 |
| chr6_90621325_90621476 | 0.16 |
Slc41a3 |
solute carrier family 41, member 3 |
2253 |
0.23 |
| chr6_72261113_72261288 | 0.16 |
Atoh8 |
atonal bHLH transcription factor 8 |
25623 |
0.12 |
| chr18_34915325_34915481 | 0.16 |
Etf1 |
eukaryotic translation termination factor 1 |
16604 |
0.11 |
| chr8_3252795_3252979 | 0.16 |
Gm16180 |
predicted gene 16180 |
6817 |
0.21 |
| chr4_61883600_61883752 | 0.16 |
Mup-ps18 |
major urinary protein, pseudogene 18 |
380 |
0.82 |
| chr9_109911713_109911882 | 0.16 |
Gm4644 |
predicted gene 4644 |
12953 |
0.11 |
| chr13_40855241_40855443 | 0.16 |
Gcnt2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
4412 |
0.12 |
| chr11_51640853_51641004 | 0.16 |
Rmnd5b |
required for meiotic nuclear division 5 homolog B |
5032 |
0.13 |
| chr8_88118582_88118749 | 0.16 |
Cnep1r1 |
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
104 |
0.95 |
| chr12_85451886_85452044 | 0.16 |
Gm40477 |
predicted gene, 40477 |
15716 |
0.14 |
| chr11_81374622_81374773 | 0.16 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
10316 |
0.3 |
| chr19_41326438_41326633 | 0.16 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
58561 |
0.12 |
| chr9_119936578_119936796 | 0.16 |
Gorasp1 |
golgi reassembly stacking protein 1 |
863 |
0.34 |
| chr6_95819185_95819380 | 0.16 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
100482 |
0.08 |
| chr7_127804573_127804884 | 0.16 |
9430064I24Rik |
RIKEN cDNA 9430064I24 gene |
1966 |
0.13 |
| chr3_27490084_27490238 | 0.16 |
Gm43344 |
predicted gene 43344 |
56480 |
0.14 |
| chr4_123527493_123527861 | 0.16 |
Macf1 |
microtubule-actin crosslinking factor 1 |
29 |
0.98 |
| chr2_33200439_33200604 | 0.16 |
Gm23546 |
predicted gene, 23546 |
11420 |
0.14 |
| chr4_129641917_129642148 | 0.16 |
Txlna |
taxilin alpha |
967 |
0.34 |
| chr14_34562012_34562194 | 0.16 |
Ldb3 |
LIM domain binding 3 |
14891 |
0.12 |
| chr4_10926160_10926369 | 0.16 |
Rps11-ps3 |
ribosomal protein S11, pseudogene 3 |
20344 |
0.15 |
| chr18_3010677_3010845 | 0.16 |
Gm50072 |
predicted gene, 50072 |
5147 |
0.22 |
| chr1_39650381_39650532 | 0.16 |
D930019O06Rik |
RIKEN cDNA D930019O06 |
280 |
0.82 |
| chr10_122384642_122385270 | 0.16 |
Gm36041 |
predicted gene, 36041 |
1936 |
0.39 |
| chr2_84386945_84387393 | 0.15 |
Calcrl |
calcitonin receptor-like |
11811 |
0.2 |
| chr8_45626518_45626714 | 0.15 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
1123 |
0.5 |
| chr8_126869593_126869763 | 0.15 |
Gm31718 |
predicted gene, 31718 |
6760 |
0.2 |
| chr17_31656771_31656922 | 0.15 |
U2af1 |
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 1 |
356 |
0.77 |
| chr8_126588770_126588952 | 0.15 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
5125 |
0.24 |
| chr16_91762844_91763094 | 0.15 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
6870 |
0.17 |
| chr1_133222741_133222940 | 0.15 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
23257 |
0.15 |
| chr9_43984454_43984635 | 0.15 |
Gm10688 |
predicted gene 10688 |
5067 |
0.11 |
| chr5_123376677_123377005 | 0.15 |
5830487J09Rik |
RIKEN cDNA 5830487J09 gene |
9842 |
0.08 |
| chr6_49166006_49166162 | 0.15 |
Gm18010 |
predicted gene, 18010 |
6675 |
0.17 |
| chr19_20698070_20698334 | 0.15 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
29360 |
0.21 |
| chr5_89018649_89018800 | 0.15 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
9242 |
0.29 |
| chr6_127254053_127254240 | 0.15 |
Gm43635 |
predicted gene 43635 |
498 |
0.71 |
| chr4_61436878_61437320 | 0.15 |
Mup15 |
major urinary protein 15 |
2644 |
0.27 |
| chr7_19956433_19956628 | 0.15 |
Rpl7a-ps8 |
ribosomal protein L7A, pseudogene 8 |
2747 |
0.12 |
| chr8_123477065_123477339 | 0.15 |
Afg3l1 |
AFG3-like AAA ATPase 1 |
701 |
0.34 |
| chr11_90252425_90252669 | 0.15 |
Mmd |
monocyte to macrophage differentiation-associated |
3071 |
0.29 |
| chr11_108782497_108782676 | 0.15 |
Cep112 |
centrosomal protein 112 |
30381 |
0.16 |
| chr4_61782137_61782580 | 0.15 |
Mup19 |
major urinary protein 19 |
89 |
0.91 |
| chr19_38223407_38224029 | 0.15 |
Fra10ac1 |
FRA10AC1 homolog (human) |
420 |
0.83 |
| chr5_73544450_73544608 | 0.15 |
Dcun1d4 |
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
33661 |
0.14 |
| chr5_89032794_89032952 | 0.15 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
4781 |
0.33 |
| chr18_24652431_24652590 | 0.15 |
Mocos |
molybdenum cofactor sulfurase |
1181 |
0.43 |
| chr16_28990306_28990479 | 0.15 |
Gm8253 |
predicted gene 8253 |
39037 |
0.19 |
| chr13_101967419_101967852 | 0.15 |
Gm17832 |
predicted gene, 17832 |
47215 |
0.17 |
| chr3_37747859_37748010 | 0.15 |
Gm42921 |
predicted gene 42921 |
13103 |
0.11 |
| chr8_22505624_22505807 | 0.15 |
Slc20a2 |
solute carrier family 20, member 2 |
410 |
0.8 |
| chr11_22276015_22276166 | 0.15 |
Ehbp1 |
EH domain binding protein 1 |
9748 |
0.28 |
| chr6_59962335_59962486 | 0.15 |
Gm18012 |
predicted gene, 18012 |
20059 |
0.25 |
| chr1_39869838_39870141 | 0.15 |
1700066B17Rik |
RIKEN cDNA 1700066B17 gene |
22655 |
0.18 |
| chrX_134248838_134248996 | 0.15 |
Nox1 |
NADPH oxidase 1 |
26961 |
0.13 |
| chr16_23135545_23135714 | 0.15 |
Gm49624 |
predicted gene, 49624 |
1309 |
0.19 |
| chr11_7197846_7198073 | 0.14 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
177 |
0.94 |
| chr6_90336860_90337033 | 0.14 |
Uroc1 |
urocanase domain containing 1 |
3657 |
0.14 |
| chr7_30640868_30641046 | 0.14 |
Rbm42 |
RNA binding motif protein 42 |
5030 |
0.07 |
| chr3_83032786_83032957 | 0.14 |
Fga |
fibrinogen alpha chain |
6656 |
0.15 |
| chr9_110702981_110703204 | 0.14 |
Ccdc12 |
coiled-coil domain containing 12 |
6802 |
0.12 |
| chr5_114556398_114556701 | 0.14 |
Gm13790 |
predicted gene 13790 |
5260 |
0.18 |
| chr6_31221431_31221835 | 0.14 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
1145 |
0.34 |
| chr8_26982042_26982193 | 0.14 |
Gm45371 |
predicted gene 45371 |
2116 |
0.17 |
| chr10_99384294_99384806 | 0.14 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
18240 |
0.17 |
| chr10_28648583_28648734 | 0.14 |
Themis |
thymocyte selection associated |
19702 |
0.24 |
| chr10_117860469_117860663 | 0.14 |
Gm40773 |
predicted gene, 40773 |
4104 |
0.17 |
| chr10_95548575_95548786 | 0.14 |
Ube2n |
ubiquitin-conjugating enzyme E2N |
7167 |
0.13 |
| chr7_29231412_29231751 | 0.14 |
Kcnk6 |
potassium inwardly-rectifying channel, subfamily K, member 6 |
274 |
0.81 |
| chr19_30092015_30092195 | 0.14 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
144 |
0.96 |
| chr4_148728491_148728680 | 0.14 |
Gm13203 |
predicted gene 13203 |
14882 |
0.18 |
| chr9_78030589_78030835 | 0.14 |
Gm47829 |
predicted gene, 47829 |
2767 |
0.16 |
| chr17_73029242_73029393 | 0.14 |
Gm30420 |
predicted gene, 30420 |
239 |
0.94 |
| chr13_92609818_92609992 | 0.14 |
Serinc5 |
serine incorporator 5 |
1186 |
0.52 |
| chr1_183349812_183350128 | 0.14 |
Mia3 |
melanoma inhibitory activity 3 |
1138 |
0.4 |
| chr11_100814569_100814933 | 0.14 |
Stat5b |
signal transducer and activator of transcription 5B |
7777 |
0.13 |
| chr10_69154715_69154964 | 0.14 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
3405 |
0.2 |
| chr4_61593182_61593358 | 0.14 |
Mup17 |
major urinary protein 17 |
2601 |
0.25 |
| chr2_179287187_179287374 | 0.14 |
Gm14293 |
predicted gene 14293 |
46790 |
0.16 |
| chr9_46217190_46217346 | 0.14 |
Sik3 |
SIK family kinase 3 |
2303 |
0.16 |
| chr15_83471603_83471756 | 0.14 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
7073 |
0.15 |
| chr12_80218314_80218489 | 0.14 |
Gm47767 |
predicted gene, 47767 |
9158 |
0.15 |
| chr1_127014601_127014752 | 0.14 |
Gm5261 |
predicted gene 5261 |
36020 |
0.21 |
| chr4_60419310_60419496 | 0.14 |
Mup9 |
major urinary protein 9 |
1181 |
0.4 |
| chr9_110335063_110335244 | 0.14 |
Scap |
SREBF chaperone |
1517 |
0.26 |
| chr2_167634687_167634863 | 0.14 |
Ube2v1 |
ubiquitin-conjugating enzyme E2 variant 1 |
2680 |
0.16 |
| chr4_132655416_132655575 | 0.14 |
Eya3 |
EYA transcriptional coactivator and phosphatase 3 |
1205 |
0.44 |
| chr8_41024281_41024448 | 0.14 |
Mtus1 |
mitochondrial tumor suppressor 1 |
1462 |
0.35 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.0 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:1901859 | negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.0 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.0 | GO:0044805 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.0 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.0 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0018630 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |