Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
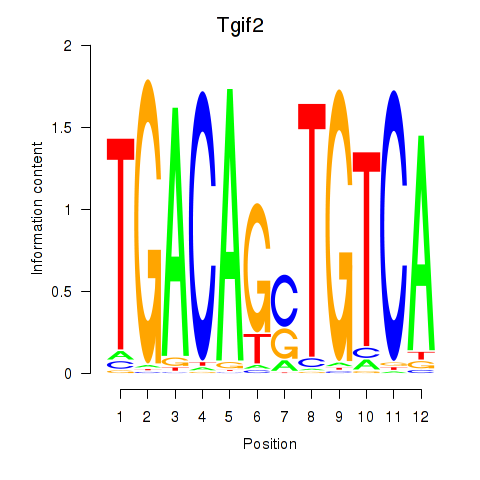
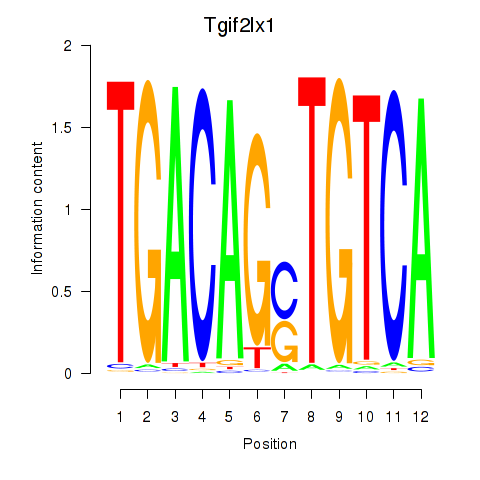
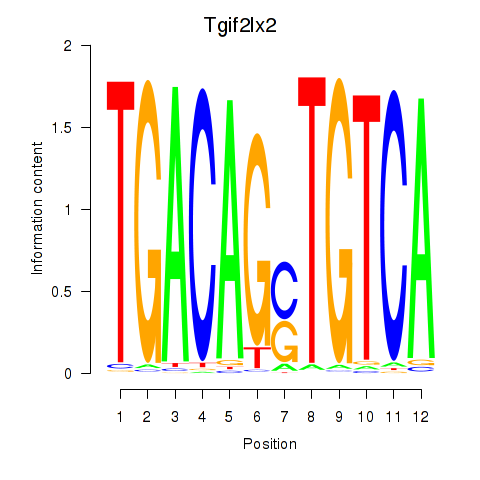
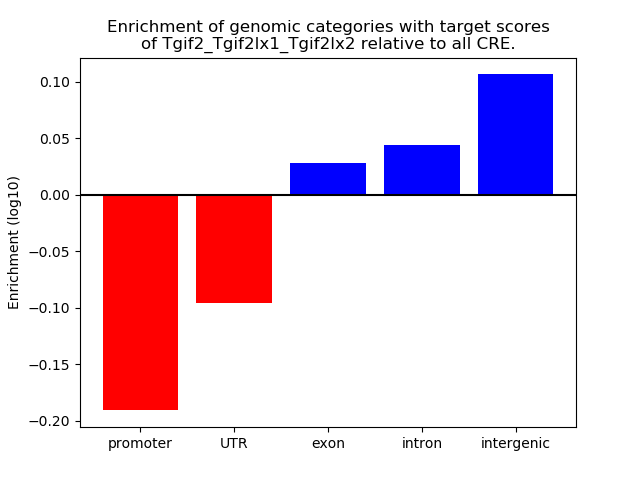
Results for Tgif2_Tgif2lx1_Tgif2lx2
Z-value: 1.12
Transcription factors associated with Tgif2_Tgif2lx1_Tgif2lx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tgif2
|
ENSMUSG00000062175.7 | TGFB-induced factor homeobox 2 |
|
Tgif2lx1
|
ENSMUSG00000061283.4 | TGFB-induced factor homeobox 2-like, X-linked 1 |
|
Tgif2lx2
|
ENSMUSG00000063242.6 | TGFB-induced factor homeobox 2-like, X-linked 2 |
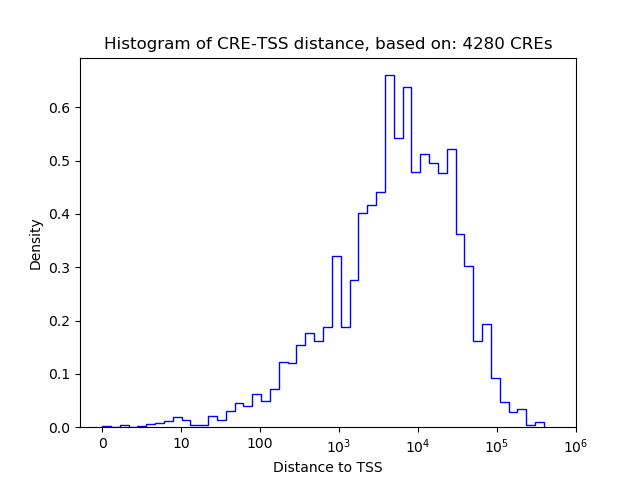
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_156841774_156841939 | Tgif2 | 834 | 0.368473 | -0.39 | 4.4e-01 | Click! |
| chr2_156850520_156850838 | Tgif2 | 157 | 0.852683 | 0.23 | 6.7e-01 | Click! |
| chr2_156842028_156842195 | Tgif2 | 1089 | 0.278758 | 0.11 | 8.3e-01 | Click! |
Activity of the Tgif2_Tgif2lx1_Tgif2lx2 motif across conditions
Conditions sorted by the z-value of the Tgif2_Tgif2lx1_Tgif2lx2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
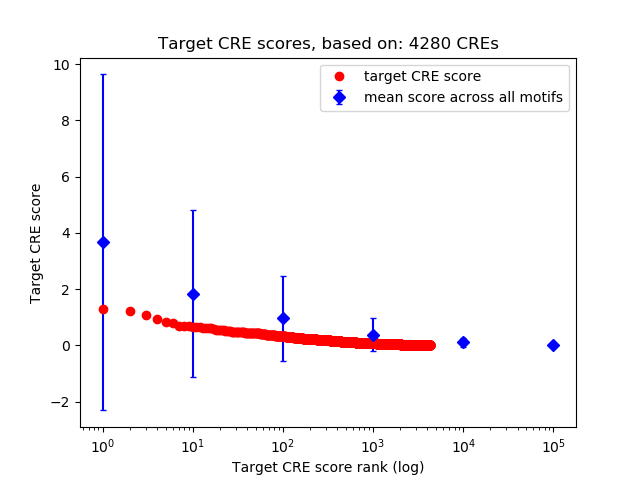
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_7200350_7200584 | 1.28 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
2685 |
0.24 |
| chr7_145087928_145088130 | 1.24 |
Gm45181 |
predicted gene 45181 |
74967 |
0.09 |
| chr19_47222639_47222810 | 1.08 |
Gm50339 |
predicted gene, 50339 |
815 |
0.48 |
| chr15_100112289_100112692 | 0.92 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
4329 |
0.17 |
| chr2_69406819_69407032 | 0.81 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
26480 |
0.18 |
| chr6_96536704_96536871 | 0.79 |
Gm26011 |
predicted gene, 26011 |
24077 |
0.25 |
| chr1_106234376_106234538 | 0.70 |
Gm38235 |
predicted gene, 38235 |
15143 |
0.2 |
| chr14_51102195_51102364 | 0.70 |
Rnase4 |
ribonuclease, RNase A family 4 |
5920 |
0.08 |
| chr13_48916144_48916308 | 0.69 |
Phf2 |
PHD finger protein 2 |
45107 |
0.13 |
| chr8_119399377_119399539 | 0.66 |
Mlycd |
malonyl-CoA decarboxylase |
4560 |
0.18 |
| chr8_123736984_123737175 | 0.66 |
Gm45781 |
predicted gene 45781 |
1937 |
0.14 |
| chr6_87428535_87428790 | 0.64 |
Bmp10 |
bone morphogenetic protein 10 |
332 |
0.85 |
| chr3_104796399_104796698 | 0.63 |
Rhoc |
ras homolog family member C |
4565 |
0.1 |
| chr9_112743581_112744166 | 0.61 |
Gm24957 |
predicted gene, 24957 |
221344 |
0.02 |
| chr5_120103213_120103364 | 0.61 |
Rbm19 |
RNA binding motif protein 19 |
13177 |
0.19 |
| chr8_119428895_119429065 | 0.60 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
5144 |
0.17 |
| chr10_95408627_95408789 | 0.59 |
Socs2 |
suppressor of cytokine signaling 2 |
4026 |
0.16 |
| chr1_9906956_9907361 | 0.55 |
Mcmdc2 |
minichromosome maintenance domain containing 2 |
1480 |
0.29 |
| chr11_69379820_69379976 | 0.54 |
Chd3 |
chromodomain helicase DNA binding protein 3 |
10492 |
0.07 |
| chr16_16225035_16225410 | 0.54 |
Pkp2 |
plakophilin 2 |
11904 |
0.19 |
| chr3_129887879_129888048 | 0.54 |
Pla2g12a |
phospholipase A2, group XIIA |
4867 |
0.17 |
| chr15_69164972_69165161 | 0.53 |
4930573C08Rik |
RIKEN cDNA 4930573C08 gene |
69933 |
0.12 |
| chr2_28525746_28525944 | 0.51 |
Ralgds |
ral guanine nucleotide dissociation stimulator |
2005 |
0.19 |
| chr2_38515308_38515472 | 0.50 |
Nek6 |
NIMA (never in mitosis gene a)-related expressed kinase 6 |
764 |
0.53 |
| chr8_126806262_126806425 | 0.50 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
32890 |
0.16 |
| chr10_81557608_81557764 | 0.49 |
Tle5 |
TLE family member 5, transcriptional modulator |
1802 |
0.15 |
| chr5_129895959_129896119 | 0.49 |
Zbed5 |
zinc finger, BED type containing 5 |
261 |
0.82 |
| chr2_167830912_167831104 | 0.48 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
2638 |
0.23 |
| chr7_58746856_58747052 | 0.48 |
Gm44937 |
predicted gene 44937 |
26755 |
0.16 |
| chr9_107667921_107668276 | 0.47 |
Slc38a3 |
solute carrier family 38, member 3 |
672 |
0.46 |
| chr8_11221370_11221555 | 0.47 |
Col4a1 |
collagen, type IV, alpha 1 |
4193 |
0.21 |
| chr2_164421410_164421570 | 0.47 |
Matn4 |
matrilin 4 |
16330 |
0.08 |
| chr14_25487639_25487838 | 0.47 |
Gm47921 |
predicted gene, 47921 |
7684 |
0.14 |
| chr3_101500478_101500642 | 0.46 |
Gm42538 |
predicted gene 42538 |
73624 |
0.07 |
| chr16_22897044_22897510 | 0.46 |
Ahsg |
alpha-2-HS-glycoprotein |
2413 |
0.17 |
| chr9_119478527_119478678 | 0.46 |
Exog |
endo/exonuclease (5'-3'), endonuclease G-like |
33615 |
0.12 |
| chr1_128705220_128705376 | 0.46 |
Gm29599 |
predicted gene 29599 |
45628 |
0.15 |
| chr8_46499145_46499431 | 0.45 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
6456 |
0.17 |
| chr1_176874134_176874434 | 0.45 |
Gm25993 |
predicted gene, 25993 |
5550 |
0.12 |
| chr11_31676998_31677176 | 0.45 |
Bod1 |
biorientation of chromosomes in cell division 1 |
5202 |
0.25 |
| chr11_98772948_98773272 | 0.45 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
2223 |
0.16 |
| chr7_35123689_35123850 | 0.44 |
Gm45091 |
predicted gene 45091 |
4164 |
0.11 |
| chr12_112561481_112561648 | 0.44 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
27220 |
0.12 |
| chr9_118101918_118102083 | 0.43 |
Azi2 |
5-azacytidine induced gene 2 |
42944 |
0.13 |
| chr6_88778293_88778444 | 0.43 |
Gm43937 |
predicted gene, 43937 |
9083 |
0.12 |
| chr4_119882590_119882770 | 0.43 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
68185 |
0.12 |
| chr3_84015637_84015957 | 0.43 |
Tmem131l |
transmembrane 131 like |
24331 |
0.2 |
| chr2_146577293_146577459 | 0.43 |
4933406D12Rik |
RIKEN cDNA 4933406D12 gene |
34445 |
0.2 |
| chr16_56063021_56063172 | 0.43 |
Gm24047 |
predicted gene, 24047 |
7467 |
0.12 |
| chr14_61045882_61046048 | 0.43 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
504 |
0.81 |
| chr1_118479315_118479504 | 0.43 |
Gm28466 |
predicted gene 28466 |
182 |
0.92 |
| chr9_75436041_75436202 | 0.42 |
Leo1 |
Leo1, Paf1/RNA polymerase II complex component |
5403 |
0.13 |
| chr2_163100058_163100224 | 0.42 |
Gm22174 |
predicted gene, 22174 |
7242 |
0.13 |
| chr14_30919586_30919927 | 0.42 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
3793 |
0.13 |
| chr5_139358617_139359046 | 0.42 |
Cyp2w1 |
cytochrome P450, family 2, subfamily w, polypeptide 1 |
4255 |
0.13 |
| chr9_25543888_25544039 | 0.42 |
Gm25861 |
predicted gene, 25861 |
4758 |
0.23 |
| chr16_80273340_80273491 | 0.41 |
Gm23083 |
predicted gene, 23083 |
136063 |
0.05 |
| chr8_68025403_68025869 | 0.41 |
Gm22018 |
predicted gene, 22018 |
15296 |
0.22 |
| chr10_93520708_93521069 | 0.41 |
Amdhd1 |
amidohydrolase domain containing 1 |
19145 |
0.12 |
| chr8_105748797_105748959 | 0.41 |
Gfod2 |
glucose-fructose oxidoreductase domain containing 2 |
9718 |
0.1 |
| chr11_5526651_5526819 | 0.40 |
Xbp1 |
X-box binding protein 1 |
4812 |
0.14 |
| chr17_73786615_73786813 | 0.39 |
Ehd3 |
EH-domain containing 3 |
17420 |
0.2 |
| chr9_61421745_61421905 | 0.39 |
Tle3 |
transducin-like enhancer of split 3 |
11888 |
0.18 |
| chr18_46993175_46993356 | 0.39 |
Gm22791 |
predicted gene, 22791 |
7784 |
0.15 |
| chr11_3182131_3182388 | 0.38 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
4766 |
0.13 |
| chr5_114982798_114982985 | 0.38 |
1810017P11Rik |
RIKEN cDNA 1810017P11 gene |
7435 |
0.09 |
| chr18_81199680_81199982 | 0.38 |
4930594M17Rik |
RIKEN cDNA 4930594M17 gene |
36235 |
0.17 |
| chr15_36902674_36902830 | 0.38 |
Gm10384 |
predicted gene 10384 |
22936 |
0.14 |
| chr6_85918490_85918716 | 0.37 |
Tprkb |
Tp53rk binding protein |
2805 |
0.1 |
| chr1_54840038_54840202 | 0.37 |
Ankrd44 |
ankyrin repeat domain 44 |
26677 |
0.18 |
| chr10_127591032_127591346 | 0.37 |
Gm16217 |
predicted gene 16217 |
5359 |
0.11 |
| chr9_78175590_78175749 | 0.37 |
C920006O11Rik |
RIKEN cDNA C920006O11 gene |
245 |
0.87 |
| chr12_86817069_86817235 | 0.36 |
Gm10095 |
predicted gene 10095 |
29315 |
0.15 |
| chr10_69288238_69288393 | 0.36 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
8857 |
0.18 |
| chr6_71235183_71235374 | 0.36 |
Smyd1 |
SET and MYND domain containing 1 |
18404 |
0.1 |
| chr19_4441349_4441500 | 0.36 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
1865 |
0.2 |
| chr7_140771105_140771348 | 0.36 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
6745 |
0.09 |
| chr17_71202252_71202409 | 0.36 |
Lpin2 |
lipin 2 |
2324 |
0.26 |
| chr18_47329689_47330033 | 0.35 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
3465 |
0.29 |
| chr15_97377850_97378467 | 0.35 |
Pced1b |
PC-esterase domain containing 1B |
16941 |
0.24 |
| chr11_118424849_118425071 | 0.35 |
C1qtnf1 |
C1q and tumor necrosis factor related protein 1 |
3243 |
0.17 |
| chr9_58295819_58296113 | 0.35 |
Loxl1 |
lysyl oxidase-like 1 |
17220 |
0.13 |
| chr2_160730308_160730465 | 0.35 |
Plcg1 |
phospholipase C, gamma 1 |
914 |
0.55 |
| chr2_169790587_169791240 | 0.35 |
Tshz2 |
teashirt zinc finger family member 2 |
93980 |
0.08 |
| chr6_125082928_125083092 | 0.34 |
2010008C14Rik |
RIKEN cDNA 2010008C14 gene |
427 |
0.62 |
| chr11_34116764_34116915 | 0.34 |
4930469K13Rik |
RIKEN cDNA 4930469K13 gene |
18285 |
0.17 |
| chr19_42690384_42690568 | 0.34 |
Gm25216 |
predicted gene, 25216 |
9426 |
0.19 |
| chr4_81673472_81673626 | 0.34 |
Gm11411 |
predicted gene 11411 |
22892 |
0.24 |
| chr8_126847749_126848310 | 0.34 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
8796 |
0.2 |
| chr4_58479203_58479397 | 0.33 |
Gm12577 |
predicted gene 12577 |
4525 |
0.22 |
| chr18_80933373_80933681 | 0.33 |
Atp9b |
ATPase, class II, type 9B |
456 |
0.78 |
| chr7_28931496_28931685 | 0.33 |
Gm44699 |
predicted gene 44699 |
11938 |
0.1 |
| chr8_10859650_10859809 | 0.33 |
Gm32540 |
predicted gene, 32540 |
6457 |
0.13 |
| chr11_82011146_82011303 | 0.32 |
Gm31522 |
predicted gene, 31522 |
8743 |
0.13 |
| chr7_140763552_140763973 | 0.32 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
23 |
0.95 |
| chr7_19864157_19864332 | 0.32 |
Ceacam16 |
carcinoembryonic antigen-related cell adhesion molecule 16 |
2945 |
0.1 |
| chr4_62393542_62393745 | 0.32 |
Slc31a1 |
solute carrier family 31, member 1 |
8142 |
0.12 |
| chr7_68696821_68697010 | 0.32 |
Gm44692 |
predicted gene 44692 |
29552 |
0.18 |
| chr1_134520125_134520290 | 0.32 |
Gm29376 |
predicted gene 29376 |
229 |
0.86 |
| chr10_84811999_84812153 | 0.32 |
Gm24226 |
predicted gene, 24226 |
893 |
0.64 |
| chr13_104060391_104060569 | 0.32 |
Nln |
neurolysin (metallopeptidase M3 family) |
1374 |
0.46 |
| chr8_13350961_13351112 | 0.32 |
Tfdp1 |
transcription factor Dp 1 |
11362 |
0.12 |
| chr7_105615844_105616248 | 0.31 |
Trim3 |
tripartite motif-containing 3 |
1980 |
0.16 |
| chr11_119219937_119220100 | 0.31 |
Gm11752 |
predicted gene 11752 |
142 |
0.92 |
| chr1_165594960_165595125 | 0.31 |
Gm38210 |
predicted gene, 38210 |
9859 |
0.11 |
| chr13_114923473_114923635 | 0.31 |
Itga2 |
integrin alpha 2 |
8546 |
0.22 |
| chr11_59640294_59640451 | 0.31 |
Mprip |
myosin phosphatase Rho interacting protein |
20933 |
0.11 |
| chr19_36731471_36731641 | 0.31 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
5097 |
0.23 |
| chr7_145044142_145044344 | 0.31 |
Ccnd1 |
cyclin D1 |
104318 |
0.05 |
| chr10_18401012_18401316 | 0.31 |
Nhsl1 |
NHS-like 1 |
6474 |
0.25 |
| chr6_43853589_43853745 | 0.31 |
Gm7783 |
predicted gene 7783 |
3237 |
0.31 |
| chr6_93758200_93758371 | 0.31 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
34254 |
0.18 |
| chr1_72790444_72790955 | 0.31 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
33804 |
0.15 |
| chr3_41582728_41583000 | 0.30 |
Jade1 |
jade family PHD finger 1 |
2008 |
0.28 |
| chr11_100322740_100322903 | 0.30 |
Eif1 |
eukaryotic translation initiation factor 1 |
1937 |
0.15 |
| chr7_137411009_137411178 | 0.30 |
9430038I01Rik |
RIKEN cDNA 9430038I01 gene |
331 |
0.89 |
| chr5_111336301_111336538 | 0.30 |
Pitpnb |
phosphatidylinositol transfer protein, beta |
5612 |
0.19 |
| chr18_84376572_84376752 | 0.30 |
Gm37216 |
predicted gene, 37216 |
516 |
0.84 |
| chr1_192392344_192392537 | 0.29 |
Kcnh1 |
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
42300 |
0.19 |
| chr2_33200439_33200604 | 0.29 |
Gm23546 |
predicted gene, 23546 |
11420 |
0.14 |
| chr9_6891499_6891736 | 0.29 |
Dync2h1 |
dynein cytoplasmic 2 heavy chain 1 |
65072 |
0.13 |
| chr2_113479270_113479457 | 0.29 |
Gm44465 |
predicted gene, 44465 |
2542 |
0.24 |
| chr9_107612485_107612643 | 0.29 |
Sema3b |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
3335 |
0.08 |
| chr4_55750335_55750505 | 0.29 |
Gm12506 |
predicted gene 12506 |
145271 |
0.04 |
| chr19_12004611_12004803 | 0.29 |
Olfr1421-ps1 |
olfactory receptor 1421, pseudogene 1 |
15468 |
0.08 |
| chr7_24487486_24487637 | 0.29 |
Cadm4 |
cell adhesion molecule 4 |
5538 |
0.09 |
| chr10_84872038_84872201 | 0.28 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
33971 |
0.16 |
| chr13_63243697_63243864 | 0.28 |
Gm47586 |
predicted gene, 47586 |
441 |
0.69 |
| chr1_34388067_34388244 | 0.28 |
Gm5266 |
predicted gene 5266 |
20784 |
0.09 |
| chr6_128481290_128481511 | 0.28 |
Pzp |
PZP, alpha-2-macroglobulin like |
6455 |
0.09 |
| chr3_138240414_138240729 | 0.28 |
0610031O16Rik |
RIKEN cDNA 0610031O16 gene |
166 |
0.88 |
| chr2_12429797_12430138 | 0.28 |
Gm13321 |
predicted gene 13321 |
7941 |
0.18 |
| chr18_60660075_60660394 | 0.28 |
Synpo |
synaptopodin |
92 |
0.97 |
| chr3_91483300_91483476 | 0.28 |
S100a7l2 |
S100 calcium binding protein A7 like 2 |
392585 |
0.01 |
| chr10_80917355_80917522 | 0.27 |
Lmnb2 |
lamin B2 |
413 |
0.7 |
| chr10_59916266_59916428 | 0.27 |
Ddit4 |
DNA-damage-inducible transcript 4 |
35487 |
0.11 |
| chr5_53437072_53437250 | 0.27 |
Gm26486 |
predicted gene, 26486 |
18588 |
0.17 |
| chr2_68873804_68874167 | 0.27 |
Cers6 |
ceramide synthase 6 |
12399 |
0.14 |
| chr7_80194026_80194200 | 0.27 |
Sema4b |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
4328 |
0.12 |
| chr11_70404879_70405056 | 0.27 |
Pelp1 |
proline, glutamic acid and leucine rich protein 1 |
5061 |
0.09 |
| chr8_105796118_105796294 | 0.27 |
Ranbp10 |
RAN binding protein 10 |
30999 |
0.07 |
| chr12_112111213_112111675 | 0.27 |
Aspg |
asparaginase |
1496 |
0.24 |
| chr13_63256313_63256480 | 0.27 |
Gm47603 |
predicted gene, 47603 |
832 |
0.41 |
| chr15_64155705_64155901 | 0.27 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
31997 |
0.18 |
| chrX_85793507_85793673 | 0.27 |
Gm6964 |
predicted gene 6964 |
530 |
0.68 |
| chrX_13062821_13063132 | 0.27 |
Usp9x |
ubiquitin specific peptidase 9, X chromosome |
8522 |
0.19 |
| chr8_83747944_83748146 | 0.26 |
Gm45778 |
predicted gene 45778 |
1459 |
0.29 |
| chr15_83524040_83524213 | 0.26 |
Bik |
BCL2-interacting killer |
2736 |
0.16 |
| chr17_42924837_42925015 | 0.26 |
Cd2ap |
CD2-associated protein |
48261 |
0.17 |
| chr16_26695984_26696378 | 0.26 |
Il1rap |
interleukin 1 receptor accessory protein |
26253 |
0.23 |
| chr17_56332362_56332517 | 0.26 |
Kdm4b |
lysine (K)-specific demethylase 4B |
6348 |
0.12 |
| chr11_74869463_74869647 | 0.26 |
Sgsm2 |
small G protein signaling modulator 2 |
463 |
0.64 |
| chr7_130248786_130248946 | 0.26 |
Fgfr2 |
fibroblast growth factor receptor 2 |
12991 |
0.27 |
| chr13_74502209_74502494 | 0.26 |
Gm49763 |
predicted gene, 49763 |
8208 |
0.1 |
| chr11_112810518_112810820 | 0.26 |
Gm11681 |
predicted gene 11681 |
11661 |
0.18 |
| chr5_112134275_112134433 | 0.26 |
1700016B01Rik |
RIKEN cDNA 1700016B01 gene |
21883 |
0.18 |
| chr19_3685828_3686010 | 0.26 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
637 |
0.59 |
| chr8_122003719_122003880 | 0.26 |
Banp |
BTG3 associated nuclear protein |
2952 |
0.19 |
| chr11_5741148_5741583 | 0.26 |
Urgcp |
upregulator of cell proliferation |
215 |
0.9 |
| chr18_81199291_81199673 | 0.25 |
4930594M17Rik |
RIKEN cDNA 4930594M17 gene |
36584 |
0.17 |
| chr5_125127560_125127724 | 0.25 |
Ncor2 |
nuclear receptor co-repressor 2 |
2742 |
0.29 |
| chr7_107633306_107633481 | 0.25 |
5330417H12Rik |
RIKEN cDNA 5330417H12 gene |
8232 |
0.16 |
| chr16_21549898_21550055 | 0.25 |
Vps8 |
VPS8 CORVET complex subunit |
13091 |
0.25 |
| chr19_16498717_16499020 | 0.25 |
Gm8222 |
predicted gene 8222 |
7804 |
0.21 |
| chr8_46332765_46332934 | 0.25 |
Gm45255 |
predicted gene 45255 |
19048 |
0.11 |
| chrX_102478577_102478728 | 0.24 |
Hdac8 |
histone deacetylase 8 |
26089 |
0.19 |
| chr5_117166861_117167023 | 0.24 |
n-R5s174 |
nuclear encoded rRNA 5S 174 |
29613 |
0.12 |
| chr11_62568400_62568605 | 0.24 |
Gm12280 |
predicted gene 12280 |
2119 |
0.14 |
| chr12_71370184_71370345 | 0.24 |
1700083H02Rik |
RIKEN cDNA 1700083H02 gene |
13944 |
0.15 |
| chr17_31930278_31930580 | 0.24 |
Gm30571 |
predicted gene, 30571 |
17763 |
0.12 |
| chr7_99141254_99141466 | 0.24 |
Uvrag |
UV radiation resistance associated gene |
219 |
0.5 |
| chr7_50868844_50869021 | 0.24 |
Gm9343 |
predicted gene 9343 |
74222 |
0.09 |
| chr5_53957420_53957607 | 0.24 |
Gm43266 |
predicted gene 43266 |
39069 |
0.14 |
| chr4_45499631_45499795 | 0.24 |
Gm22518 |
predicted gene, 22518 |
839 |
0.56 |
| chr5_32735922_32736077 | 0.24 |
Gm43695 |
predicted gene 43695 |
3335 |
0.14 |
| chr3_94310992_94311165 | 0.24 |
Them4 |
thioesterase superfamily member 4 |
942 |
0.35 |
| chr5_130761960_130762123 | 0.24 |
Gm23761 |
predicted gene, 23761 |
20660 |
0.24 |
| chr1_155886750_155887199 | 0.24 |
Cep350 |
centrosomal protein 350 |
960 |
0.44 |
| chr14_65100574_65100746 | 0.24 |
Extl3 |
exostosin-like glycosyltransferase 3 |
2554 |
0.29 |
| chr17_79904674_79904846 | 0.24 |
Atl2 |
atlastin GTPase 2 |
8637 |
0.18 |
| chr5_92773632_92773840 | 0.24 |
Mir1961 |
microRNA 1961 |
14826 |
0.18 |
| chr16_21642073_21642273 | 0.24 |
Vps8 |
VPS8 CORVET complex subunit |
14542 |
0.21 |
| chr4_134931334_134931504 | 0.23 |
Syf2 |
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
409 |
0.81 |
| chr11_45145769_45145947 | 0.23 |
Gm12160 |
predicted gene 12160 |
15085 |
0.2 |
| chr13_33935183_33935334 | 0.23 |
1110046J04Rik |
RIKEN cDNA 1110046J04 gene |
309 |
0.67 |
| chr16_28989883_28990229 | 0.23 |
Gm8253 |
predicted gene 8253 |
38701 |
0.19 |
| chr2_59193239_59193408 | 0.23 |
Pkp4 |
plakophilin 4 |
7451 |
0.22 |
| chr4_82621551_82621752 | 0.23 |
Gm11268 |
predicted gene 11268 |
20749 |
0.19 |
| chr9_70672050_70672218 | 0.23 |
Adam10 |
a disintegrin and metallopeptidase domain 10 |
6863 |
0.18 |
| chr5_114772969_114773120 | 0.23 |
Git2 |
GIT ArfGAP 2 |
390 |
0.68 |
| chr3_18080789_18080940 | 0.23 |
Gm23726 |
predicted gene, 23726 |
11538 |
0.2 |
| chr4_149887083_149887262 | 0.23 |
Gm13070 |
predicted gene 13070 |
16834 |
0.13 |
| chr2_93823530_93823821 | 0.23 |
Ext2 |
exostosin glycosyltransferase 2 |
1107 |
0.46 |
| chr8_115089778_115090403 | 0.23 |
Gm22556 |
predicted gene, 22556 |
37177 |
0.23 |
| chr9_48592438_48592815 | 0.23 |
Nnmt |
nicotinamide N-methyltransferase |
4749 |
0.26 |
| chr1_40221808_40222119 | 0.22 |
Il1r1 |
interleukin 1 receptor, type I |
3117 |
0.26 |
| chr15_27800508_27800681 | 0.22 |
Trio |
triple functional domain (PTPRF interacting) |
11956 |
0.22 |
| chr3_57435613_57435790 | 0.22 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
10387 |
0.22 |
| chr7_48883645_48884044 | 0.22 |
E2f8 |
E2F transcription factor 8 |
2248 |
0.2 |
| chr4_149098057_149098222 | 0.22 |
Pex14 |
peroxisomal biogenesis factor 14 |
1670 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.3 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.2 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.2 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0071673 | regulation of smooth muscle cell chemotaxis(GO:0071671) positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.0 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.0 | 0.2 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0014744 | positive regulation of muscle adaptation(GO:0014744) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.3 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0018655 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.0 | GO:0080084 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |