Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
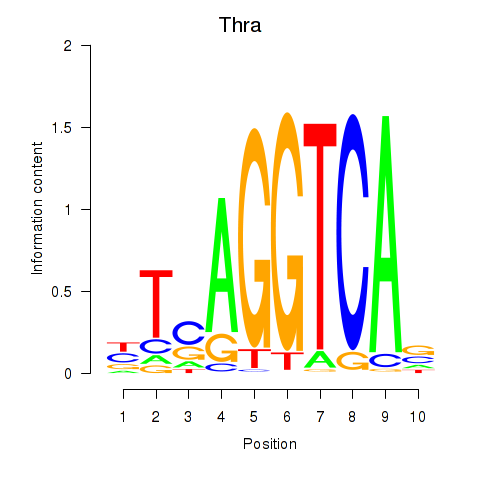
Results for Thra
Z-value: 2.01
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSMUSG00000058756.7 | thyroid hormone receptor alpha |
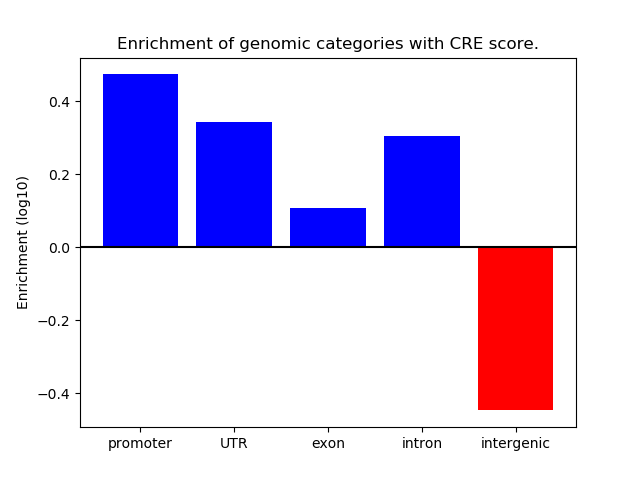
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_98740979_98741208 | Thra | 189 | 0.886840 | -0.96 | 2.9e-03 | Click! |
| chr11_98742155_98742392 | Thra | 115 | 0.927284 | -0.88 | 2.2e-02 | Click! |
| chr11_98741745_98741898 | Thra | 10 | 0.952548 | -0.87 | 2.3e-02 | Click! |
| chr11_98754405_98754602 | Thra | 917 | 0.389186 | -0.83 | 4.0e-02 | Click! |
| chr11_98747337_98747676 | Thra | 3273 | 0.126132 | 0.77 | 7.1e-02 | Click! |
Activity of the Thra motif across conditions
Conditions sorted by the z-value of the Thra motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
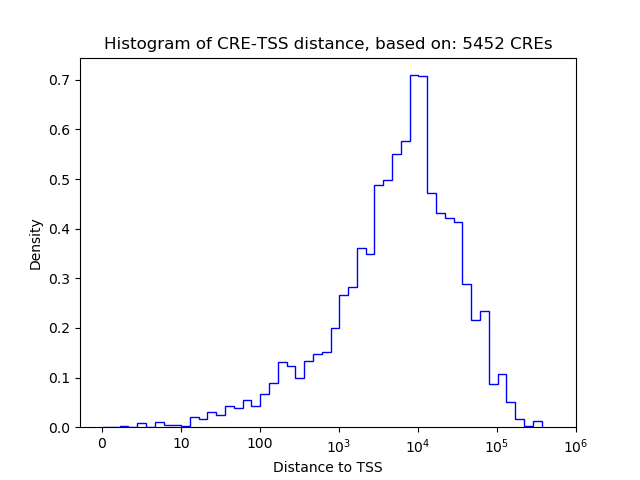
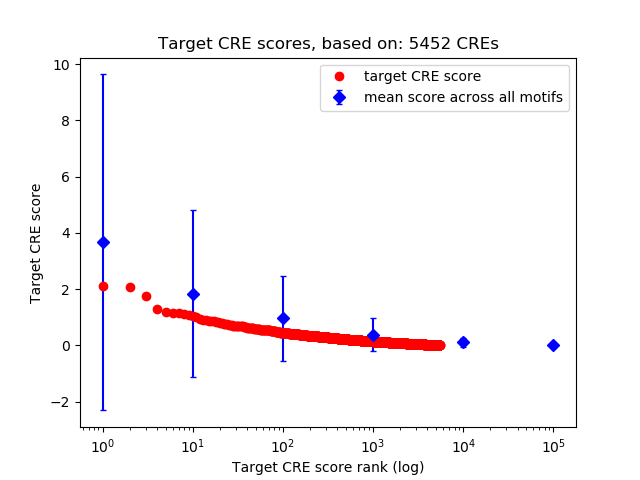
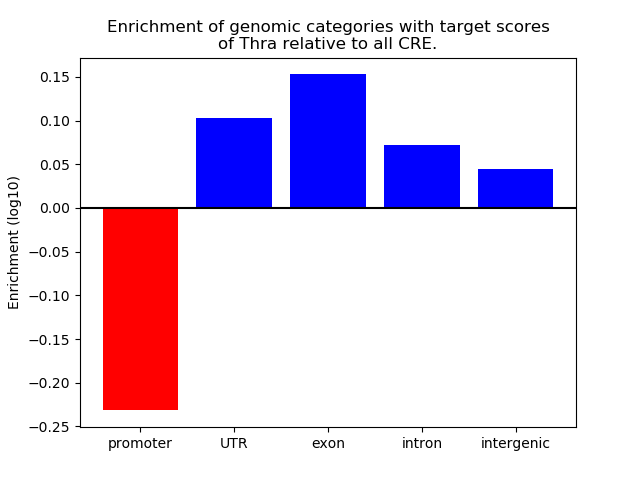
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_149579767_149579923 | 2.10 |
Wdr95 |
WD40 repeat domain 95 |
1933 |
0.28 |
| chr1_193928644_193928965 | 2.06 |
Gm21362 |
predicted gene, 21362 |
61795 |
0.15 |
| chr6_136954641_136954792 | 1.75 |
Pde6h |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
183 |
0.93 |
| chr10_89813381_89813532 | 1.31 |
Uhrf1bp1l |
UHRF1 (ICBP90) binding protein 1-like |
1585 |
0.4 |
| chr9_74523907_74524058 | 1.19 |
Gm28622 |
predicted gene 28622 |
37396 |
0.17 |
| chr11_16862611_16863177 | 1.17 |
Egfr |
epidermal growth factor receptor |
15256 |
0.19 |
| chr2_33224574_33224733 | 1.16 |
Angptl2 |
angiopoietin-like 2 |
8536 |
0.16 |
| chr4_128946455_128946665 | 1.11 |
Gm15904 |
predicted gene 15904 |
10555 |
0.15 |
| chr7_26313878_26314034 | 1.06 |
Gm42375 |
predicted gene, 42375 |
6670 |
0.14 |
| chrX_85734354_85734533 | 1.05 |
Gk |
glycerol kinase |
3167 |
0.2 |
| chr3_107238408_107238575 | 1.01 |
Prok1 |
prokineticin 1 |
161 |
0.93 |
| chr10_87931882_87932033 | 0.95 |
Tyms-ps |
thymidylate synthase, pseudogene |
34890 |
0.14 |
| chr18_79035328_79035493 | 0.89 |
Setbp1 |
SET binding protein 1 |
73981 |
0.12 |
| chr2_58766481_58766674 | 0.89 |
Upp2 |
uridine phosphorylase 2 |
1252 |
0.49 |
| chr4_136689035_136689208 | 0.87 |
Ephb2 |
Eph receptor B2 |
4919 |
0.19 |
| chr3_64420614_64420765 | 0.87 |
Vmn2r4 |
vomeronasal 2, receptor 4 |
5321 |
0.14 |
| chr4_150660019_150660333 | 0.86 |
Slc45a1 |
solute carrier family 45, member 1 |
8002 |
0.19 |
| chr13_107641777_107642027 | 0.84 |
Gm32090 |
predicted gene, 32090 |
29053 |
0.18 |
| chr15_84115082_84115482 | 0.83 |
Pnpla5 |
patatin-like phospholipase domain containing 5 |
7893 |
0.11 |
| chr18_55182860_55183019 | 0.80 |
Gm22597 |
predicted gene, 22597 |
3099 |
0.29 |
| chr19_12720308_12720459 | 0.78 |
Gm15962 |
predicted gene 15962 |
820 |
0.46 |
| chr5_110790735_110790921 | 0.77 |
Ulk1 |
unc-51 like kinase 1 |
1362 |
0.28 |
| chr10_62647927_62648078 | 0.76 |
Ddx50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
221 |
0.89 |
| chr5_65999169_65999357 | 0.75 |
9130230L23Rik |
RIKEN cDNA 9130230L23 gene |
955 |
0.42 |
| chr19_32905404_32905569 | 0.73 |
Gm36860 |
predicted gene, 36860 |
74577 |
0.1 |
| chr8_66494561_66494830 | 0.72 |
Gm32568 |
predicted gene, 32568 |
364 |
0.85 |
| chr6_121170717_121171144 | 0.72 |
Pex26 |
peroxisomal biogenesis factor 26 |
12737 |
0.13 |
| chr2_9014159_9014316 | 0.70 |
Gm13217 |
predicted gene 13217 |
28247 |
0.25 |
| chr13_113844856_113845031 | 0.69 |
Arl15 |
ADP-ribosylation factor-like 15 |
50321 |
0.11 |
| chr8_122465127_122465303 | 0.69 |
Rnf166 |
ring finger protein 166 |
3012 |
0.11 |
| chr4_135649660_135649811 | 0.68 |
1700029M20Rik |
RIKEN cDNA 1700029M20 gene |
22482 |
0.13 |
| chr2_20909493_20909665 | 0.68 |
Arhgap21 |
Rho GTPase activating protein 21 |
5115 |
0.23 |
| chr1_169165587_169165766 | 0.68 |
Mir6354 |
microRNA 6354 |
146587 |
0.04 |
| chr4_124005727_124005878 | 0.68 |
Gm12902 |
predicted gene 12902 |
79568 |
0.07 |
| chr7_81684160_81684325 | 0.67 |
Homer2 |
homer scaffolding protein 2 |
8645 |
0.14 |
| chr11_60205641_60205948 | 0.67 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
318 |
0.81 |
| chr8_9989223_9989374 | 0.66 |
Abhd13 |
abhydrolase domain containing 13 |
11581 |
0.14 |
| chr19_18638919_18639112 | 0.65 |
Nmrk1 |
nicotinamide riboside kinase 1 |
7017 |
0.17 |
| chr4_139219800_139219985 | 0.63 |
Capzb |
capping protein (actin filament) muscle Z-line, beta |
7069 |
0.16 |
| chr1_156813617_156813768 | 0.63 |
Ralgps2 |
Ral GEF with PH domain and SH3 binding motif 2 |
124 |
0.95 |
| chr8_114150614_114151007 | 0.63 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
17168 |
0.25 |
| chr10_66912996_66913163 | 0.62 |
1110002J07Rik |
RIKEN cDNA 1110002J07 gene |
4660 |
0.18 |
| chr2_29627943_29628094 | 0.62 |
Rapgef1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
8108 |
0.21 |
| chr13_73367970_73368135 | 0.62 |
Eprn |
ephemeron, early developmental lncRNA |
20668 |
0.15 |
| chr9_22493039_22493190 | 0.61 |
Bbs9 |
Bardet-Biedl syndrome 9 (human) |
2285 |
0.22 |
| chr6_118939696_118939861 | 0.61 |
Gm25905 |
predicted gene, 25905 |
104878 |
0.07 |
| chr9_73084122_73084303 | 0.61 |
Rab27a |
RAB27A, member RAS oncogene family |
10621 |
0.09 |
| chr5_101949474_101949645 | 0.60 |
Gm42934 |
predicted gene 42934 |
7048 |
0.21 |
| chr3_133739026_133739177 | 0.59 |
Gm6135 |
prediticted gene 6135 |
52403 |
0.13 |
| chr4_131722484_131723038 | 0.58 |
Gm16080 |
predicted gene 16080 |
12253 |
0.19 |
| chr17_31324627_31325122 | 0.58 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
753 |
0.58 |
| chr4_11974538_11974711 | 0.58 |
1700123M08Rik |
RIKEN cDNA 1700123M08 gene |
5538 |
0.16 |
| chrX_52033079_52033272 | 0.57 |
1700080O16Rik |
RIKEN cDNA 1700080O16 gene |
60403 |
0.14 |
| chr3_122324623_122324795 | 0.57 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
30573 |
0.09 |
| chr7_81685163_81685588 | 0.57 |
Homer2 |
homer scaffolding protein 2 |
7512 |
0.14 |
| chr11_118107158_118107309 | 0.56 |
Gm11738 |
predicted gene 11738 |
7622 |
0.17 |
| chr3_152091714_152091865 | 0.56 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
11065 |
0.14 |
| chr6_22198707_22198884 | 0.56 |
Gm42573 |
predicted gene 42573 |
47139 |
0.15 |
| chr12_16585344_16585551 | 0.56 |
Lpin1 |
lipin 1 |
4273 |
0.27 |
| chr1_133236046_133236205 | 0.55 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
9972 |
0.16 |
| chr8_24455254_24455437 | 0.55 |
Gm44620 |
predicted gene 44620 |
8248 |
0.15 |
| chr11_60203795_60204262 | 0.54 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
171 |
0.9 |
| chrX_51390004_51390155 | 0.54 |
Gm14621 |
predicted gene 14621 |
150547 |
0.04 |
| chr12_80932956_80933128 | 0.54 |
1700052I22Rik |
RIKEN cDNA 1700052I22 gene |
8593 |
0.13 |
| chr2_181276195_181276365 | 0.54 |
Gmeb2 |
glucocorticoid modulatory element binding protein 2 |
1802 |
0.2 |
| chr8_12772445_12772606 | 0.53 |
Atp11a |
ATPase, class VI, type 11A |
15456 |
0.16 |
| chr17_78209009_78209169 | 0.53 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
8024 |
0.15 |
| chr17_14399074_14399237 | 0.53 |
Smoc2 |
SPARC related modular calcium binding 2 |
85 |
0.68 |
| chr14_55019593_55019773 | 0.53 |
Ngdn |
neuroguidin, EIF4E binding protein |
1206 |
0.23 |
| chr7_26951142_26951507 | 0.53 |
Cyp2a22 |
cytochrome P450, family 2, subfamily a, polypeptide 22 |
11940 |
0.14 |
| chr17_34855282_34855491 | 0.53 |
Nelfe |
negative elongation factor complex member E, Rdbp |
1130 |
0.17 |
| chr19_37074413_37074612 | 0.53 |
Gm22714 |
predicted gene, 22714 |
74650 |
0.08 |
| chr11_16874000_16874630 | 0.53 |
Egfr |
epidermal growth factor receptor |
3835 |
0.25 |
| chr6_70874727_70874899 | 0.52 |
Eif2ak3 |
eukaryotic translation initiation factor 2 alpha kinase 3 |
3748 |
0.18 |
| chr18_81321681_81321954 | 0.52 |
Gm30192 |
predicted gene, 30192 |
57109 |
0.13 |
| chr1_16235159_16235461 | 0.52 |
Gm28095 |
predicted gene 28095 |
6804 |
0.19 |
| chr17_23535025_23535512 | 0.52 |
6330415G19Rik |
RIKEN cDNA 6330415G19 gene |
15531 |
0.08 |
| chr11_16842103_16842406 | 0.52 |
Egfros |
epidermal growth factor receptor, opposite strand |
11552 |
0.21 |
| chr9_72385313_72385484 | 0.51 |
Gm37150 |
predicted gene, 37150 |
1009 |
0.31 |
| chrX_52047104_52047283 | 0.50 |
1700080O16Rik |
RIKEN cDNA 1700080O16 gene |
74421 |
0.11 |
| chr9_74894770_74895613 | 0.50 |
Onecut1 |
one cut domain, family member 1 |
28707 |
0.13 |
| chr15_84113983_84114148 | 0.49 |
Gm46526 |
predicted gene, 46526 |
7407 |
0.11 |
| chr2_172973899_172974057 | 0.49 |
Spo11 |
SPO11 meiotic protein covalently bound to DSB |
3722 |
0.19 |
| chr11_16836874_16837160 | 0.49 |
Egfros |
epidermal growth factor receptor, opposite strand |
6315 |
0.23 |
| chr3_18148711_18148862 | 0.48 |
Gm23686 |
predicted gene, 23686 |
28839 |
0.19 |
| chr2_58565325_58565706 | 0.47 |
Acvr1 |
activin A receptor, type 1 |
1311 |
0.41 |
| chr1_91618886_91619039 | 0.47 |
Gm28380 |
predicted gene 28380 |
7556 |
0.22 |
| chr13_62868476_62868715 | 0.47 |
Fbp1 |
fructose bisphosphatase 1 |
4163 |
0.15 |
| chr1_153739671_153739993 | 0.47 |
Rgs16 |
regulator of G-protein signaling 16 |
517 |
0.61 |
| chr17_71250166_71250322 | 0.46 |
Lpin2 |
lipin 2 |
4213 |
0.18 |
| chr4_65080602_65080759 | 0.46 |
Pappa |
pregnancy-associated plasma protein A |
43494 |
0.19 |
| chr4_53201741_53202070 | 0.46 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
15656 |
0.18 |
| chr4_121078016_121078199 | 0.46 |
Zmpste24 |
zinc metallopeptidase, STE24 |
369 |
0.74 |
| chr5_104031868_104032176 | 0.46 |
Hsd17b11 |
hydroxysteroid (17-beta) dehydrogenase 11 |
10103 |
0.11 |
| chr18_68491528_68491858 | 0.45 |
Gm50258 |
predicted gene, 50258 |
58900 |
0.1 |
| chr5_66012709_66012860 | 0.45 |
9130230L23Rik |
RIKEN cDNA 9130230L23 gene |
8499 |
0.12 |
| chr3_100892764_100892976 | 0.45 |
Trim45 |
tripartite motif-containing 45 |
29332 |
0.16 |
| chr5_130009895_130010078 | 0.44 |
Asl |
argininosuccinate lyase |
4736 |
0.13 |
| chr5_32155224_32155404 | 0.44 |
Fosl2 |
fos-like antigen 2 |
8793 |
0.15 |
| chr13_44291297_44291488 | 0.44 |
Gm29676 |
predicted gene, 29676 |
10943 |
0.21 |
| chr1_65383811_65383972 | 0.44 |
Gm28927 |
predicted gene 28927 |
37230 |
0.11 |
| chr11_71018794_71019067 | 0.44 |
Derl2 |
Der1-like domain family, member 2 |
282 |
0.7 |
| chr9_121118120_121118389 | 0.44 |
Ulk4 |
unc-51-like kinase 4 |
9729 |
0.23 |
| chr3_133517548_133517726 | 0.44 |
Tet2 |
tet methylcytosine dioxygenase 2 |
26649 |
0.15 |
| chr5_115585229_115585380 | 0.44 |
Gcn1 |
GCN1 activator of EIF2AK4 |
2907 |
0.14 |
| chr6_35335576_35335734 | 0.43 |
1700065J11Rik |
RIKEN cDNA 1700065J11 gene |
4809 |
0.19 |
| chr3_89133157_89133825 | 0.43 |
Pklr |
pyruvate kinase liver and red blood cell |
2651 |
0.11 |
| chr12_112591206_112591385 | 0.43 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
2029 |
0.25 |
| chr14_24960151_24960328 | 0.43 |
Gm10398 |
predicted gene 10398 |
50825 |
0.13 |
| chr7_97423614_97423975 | 0.43 |
Thrsp |
thyroid hormone responsive |
6064 |
0.14 |
| chr19_40228198_40228457 | 0.43 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
2478 |
0.23 |
| chr9_106725564_106725762 | 0.43 |
Rad54l2 |
RAD54 like 2 (S. cerevisiae) |
2792 |
0.23 |
| chr3_145591903_145592058 | 0.42 |
Znhit6 |
zinc finger, HIT type 6 |
3888 |
0.24 |
| chr15_84156345_84156653 | 0.42 |
Pnpla3 |
patatin-like phospholipase domain containing 3 |
11338 |
0.1 |
| chr15_85233381_85233563 | 0.42 |
Fbln1 |
fibulin 1 |
27467 |
0.17 |
| chr11_16799464_16799909 | 0.42 |
Egfros |
epidermal growth factor receptor, opposite strand |
31016 |
0.16 |
| chr11_118399939_118400090 | 0.42 |
Lgals3bp |
lectin, galactoside-binding, soluble, 3 binding protein |
1024 |
0.44 |
| chr17_55512009_55512180 | 0.42 |
St6gal2 |
beta galactoside alpha 2,6 sialyltransferase 2 |
65850 |
0.11 |
| chr7_102031157_102031308 | 0.42 |
Rnf121 |
ring finger protein 121 |
6763 |
0.1 |
| chr16_97511614_97511792 | 0.42 |
Fam3b |
family with sequence similarity 3, member B |
827 |
0.52 |
| chr5_124094116_124094363 | 0.42 |
Abcb9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
1547 |
0.24 |
| chr7_44684146_44684300 | 0.41 |
2310016G11Rik |
RIKEN cDNA 2310016G11 gene |
7369 |
0.1 |
| chr3_100167851_100168005 | 0.41 |
Gdap2 |
ganglioside-induced differentiation-associated-protein 2 |
5384 |
0.21 |
| chr19_21624664_21624891 | 0.41 |
1110059E24Rik |
RIKEN cDNA 1110059E24 gene |
5991 |
0.23 |
| chr1_163338521_163338892 | 0.41 |
Prrx1 |
paired related homeobox 1 |
24996 |
0.16 |
| chr5_33605422_33605612 | 0.41 |
Fam53a |
family with sequence similarity 53, member A |
23395 |
0.11 |
| chr2_4895477_4895652 | 0.41 |
Sephs1 |
selenophosphate synthetase 1 |
11180 |
0.15 |
| chrX_88826455_88826619 | 0.41 |
Gm27000 |
predicted gene, 27000 |
66225 |
0.08 |
| chr6_52007436_52007714 | 0.41 |
Skap2 |
src family associated phosphoprotein 2 |
4940 |
0.18 |
| chr4_45342370_45342984 | 0.41 |
Exosc3 |
exosome component 3 |
55 |
0.88 |
| chr19_8097849_8098000 | 0.40 |
Slc22a28 |
solute carrier family 22, member 28 |
34058 |
0.16 |
| chr6_35338820_35339318 | 0.40 |
1700065J11Rik |
RIKEN cDNA 1700065J11 gene |
8223 |
0.18 |
| chr9_56173489_56173658 | 0.40 |
Tspan3 |
tetraspanin 3 |
8096 |
0.17 |
| chr2_172684895_172685060 | 0.40 |
Tfap2c |
transcription factor AP-2, gamma |
132861 |
0.04 |
| chr4_111200292_111200482 | 0.40 |
Agbl4 |
ATP/GTP binding protein-like 4 |
81476 |
0.1 |
| chr5_125178269_125178781 | 0.40 |
Ncor2 |
nuclear receptor co-repressor 2 |
528 |
0.81 |
| chr11_22493852_22494015 | 0.40 |
Gm12051 |
predicted gene 12051 |
10325 |
0.19 |
| chr6_124674733_124674894 | 0.39 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
11598 |
0.07 |
| chr5_115345253_115345441 | 0.39 |
Cox6a1 |
cytochrome c oxidase subunit 6A1 |
3567 |
0.09 |
| chr2_93755436_93755602 | 0.39 |
Gm13818 |
predicted gene 13818 |
39051 |
0.14 |
| chr5_99423905_99424056 | 0.39 |
Gm35172 |
predicted gene, 35172 |
84409 |
0.08 |
| chr6_145855716_145856207 | 0.39 |
Gm43909 |
predicted gene, 43909 |
7336 |
0.17 |
| chr12_101990840_101991003 | 0.39 |
Ndufb1-ps |
NADH:ubiquinone oxidoreductase subunit B1 |
13853 |
0.15 |
| chr3_129444515_129444714 | 0.39 |
Gm5712 |
predicted gene 5712 |
10941 |
0.17 |
| chr11_51591243_51591573 | 0.39 |
Phykpl |
5-phosphohydroxy-L-lysine phospholyase |
39 |
0.96 |
| chr10_86828681_86828832 | 0.39 |
Nt5dc3 |
5'-nucleotidase domain containing 3 |
3653 |
0.16 |
| chr8_56311225_56311376 | 0.39 |
Gm45540 |
predicted gene 45540 |
4 |
0.98 |
| chr13_75413535_75413744 | 0.39 |
Gm48234 |
predicted gene, 48234 |
65748 |
0.11 |
| chr3_104674294_104674500 | 0.39 |
Gm29560 |
predicted gene 29560 |
4387 |
0.11 |
| chr16_43545329_43545492 | 0.39 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
35102 |
0.18 |
| chr7_112763652_112763828 | 0.38 |
Tead1 |
TEA domain family member 1 |
4194 |
0.26 |
| chr5_91293069_91293447 | 0.38 |
Gm19619 |
predicted gene, 19619 |
9837 |
0.24 |
| chr9_108404599_108404954 | 0.38 |
1700102P08Rik |
RIKEN cDNA 1700102P08 gene |
11942 |
0.07 |
| chr17_66041118_66041288 | 0.38 |
Ankrd12 |
ankyrin repeat domain 12 |
35261 |
0.1 |
| chr11_16815536_16815687 | 0.38 |
Egfros |
epidermal growth factor receptor, opposite strand |
15091 |
0.21 |
| chr2_107127395_107127560 | 0.38 |
Gm13903 |
predicted gene 13903 |
3517 |
0.26 |
| chr4_150230846_150231004 | 0.38 |
Gm13094 |
predicted gene 13094 |
2890 |
0.19 |
| chr1_172373715_172373866 | 0.38 |
Pigm |
phosphatidylinositol glycan anchor biosynthesis, class M |
2756 |
0.17 |
| chr2_155067847_155068055 | 0.38 |
Gm45609 |
predicted gene 45609 |
6230 |
0.14 |
| chr4_149150340_149150711 | 0.38 |
Rpsa-ps12 |
ribosomal protein SA, pseudogene 12 |
7137 |
0.12 |
| chr14_33933061_33933242 | 0.38 |
Gdf2 |
growth differentiation factor 2 |
7888 |
0.14 |
| chr18_67693755_67693945 | 0.37 |
Ptpn2 |
protein tyrosine phosphatase, non-receptor type 2 |
3626 |
0.22 |
| chr7_123643560_123643752 | 0.37 |
Zkscan2 |
zinc finger with KRAB and SCAN domains 2 |
143207 |
0.04 |
| chr18_79086140_79086307 | 0.37 |
Setbp1 |
SET binding protein 1 |
23168 |
0.26 |
| chr19_7309675_7309826 | 0.37 |
Gm17227 |
predicted gene 17227 |
12016 |
0.11 |
| chr4_137896248_137896586 | 0.36 |
Gm13012 |
predicted gene 13012 |
12480 |
0.19 |
| chr2_52319247_52319421 | 0.36 |
Neb |
nebulin |
15800 |
0.17 |
| chr13_50011845_50011996 | 0.36 |
Gm6554 |
predicted gene 6554 |
9662 |
0.13 |
| chr2_118399841_118399992 | 0.36 |
Eif2ak4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
10804 |
0.16 |
| chr14_64139029_64139418 | 0.36 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
22909 |
0.13 |
| chr5_143053277_143053452 | 0.36 |
Gm43378 |
predicted gene 43378 |
3321 |
0.18 |
| chr8_76928418_76928588 | 0.36 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
20231 |
0.2 |
| chr9_44289013_44289164 | 0.36 |
Abcg4 |
ATP binding cassette subfamily G member 4 |
473 |
0.55 |
| chr8_128479257_128479434 | 0.36 |
Nrp1 |
neuropilin 1 |
119948 |
0.05 |
| chr11_5505466_5505686 | 0.36 |
Gm11963 |
predicted gene 11963 |
6736 |
0.14 |
| chr6_144707085_144707314 | 0.36 |
Sox5os4 |
SRY (sex determining region Y)-box 5, opposite strand 4 |
13712 |
0.16 |
| chr11_23148992_23149143 | 0.35 |
1700061J23Rik |
RIKEN cDNA 1700061J23 gene |
3073 |
0.24 |
| chr4_15937312_15937486 | 0.35 |
Mir6400 |
microRNA 6400 |
5592 |
0.13 |
| chr17_23533858_23534146 | 0.35 |
6330415G19Rik |
RIKEN cDNA 6330415G19 gene |
16797 |
0.08 |
| chr13_50776099_50776250 | 0.35 |
Gm2654 |
predicted gene 2654 |
9970 |
0.19 |
| chr8_108786077_108786260 | 0.35 |
Gm38042 |
predicted gene, 38042 |
48575 |
0.15 |
| chr6_145857491_145857666 | 0.35 |
Gm43909 |
predicted gene, 43909 |
5719 |
0.18 |
| chr10_121369241_121369437 | 0.35 |
Gns |
glucosamine (N-acetyl)-6-sulfatase |
4163 |
0.15 |
| chr7_71232680_71232844 | 0.35 |
6030442E23Rik |
RIKEN cDNA 6030442E23 gene |
15713 |
0.2 |
| chr19_43747913_43748064 | 0.35 |
Cutc |
cutC copper transporter |
5008 |
0.14 |
| chr12_76927037_76927196 | 0.35 |
Max |
Max protein |
12878 |
0.15 |
| chr1_67148058_67148242 | 0.35 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
25124 |
0.21 |
| chr1_134564053_134564250 | 0.35 |
Kdm5b |
lysine (K)-specific demethylase 5B |
3944 |
0.16 |
| chr11_21082776_21082975 | 0.35 |
Peli1 |
pellino 1 |
8416 |
0.2 |
| chr7_107700018_107700215 | 0.35 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
34805 |
0.11 |
| chr15_69062103_69062352 | 0.35 |
Gm49422 |
predicted gene, 49422 |
102696 |
0.07 |
| chr15_89477372_89477550 | 0.34 |
Arsa |
arylsulfatase A |
36 |
0.77 |
| chr4_139494245_139494414 | 0.34 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
13949 |
0.17 |
| chr8_4616055_4616227 | 0.34 |
Zfp958 |
zinc finger protein 958 |
2974 |
0.19 |
| chr3_18161234_18161612 | 0.34 |
Gm23686 |
predicted gene, 23686 |
16202 |
0.22 |
| chr8_115830386_115830552 | 0.34 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
122675 |
0.06 |
| chr13_9810187_9810338 | 0.34 |
Gm47401 |
predicted gene, 47401 |
10249 |
0.16 |
| chr15_38103232_38103407 | 0.34 |
Gm49312 |
predicted gene, 49312 |
24217 |
0.14 |
| chr11_60200538_60200855 | 0.34 |
Mir6921 |
microRNA 6921 |
79 |
0.79 |
| chr11_106490027_106490381 | 0.34 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
2352 |
0.23 |
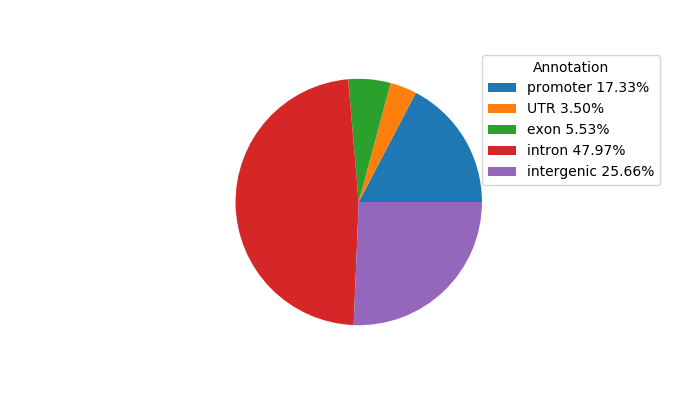
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.4 | 1.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.6 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.6 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.1 | 0.3 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.2 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 1.7 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.2 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.4 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.4 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.4 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.2 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0035935 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:1903802 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:0035482 | gastric motility(GO:0035482) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0042504 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.2 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.0 | 0.0 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.0 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.2 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) |
| 0.0 | 0.1 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.0 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.0 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.0 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.0 | 0.0 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.2 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0048818 | positive regulation of hair follicle maturation(GO:0048818) |
| 0.0 | 0.0 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 0.0 | 0.1 | GO:0060337 | type I interferon signaling pathway(GO:0060337) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.0 | 0.0 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.0 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.3 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.2 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0052760 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | Genes involved in NFkB and MAP kinases activation mediated by TLR4 signaling repertoire |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |