Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
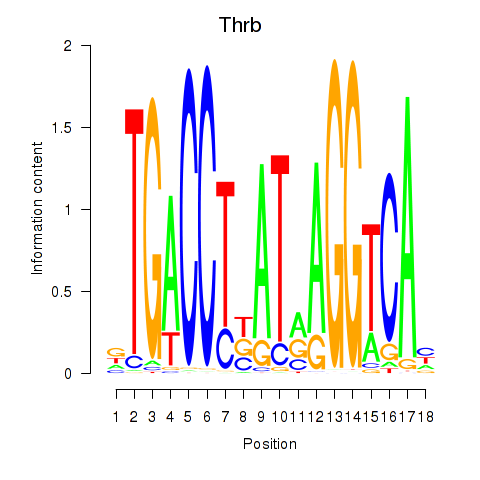
Results for Thrb
Z-value: 1.82
Transcription factors associated with Thrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thrb
|
ENSMUSG00000021779.10 | thyroid hormone receptor beta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_17695776_17695955 | Thrb | 34969 | 0.214039 | -0.98 | 8.9e-04 | Click! |
| chr14_17681136_17681362 | Thrb | 20353 | 0.265627 | -0.95 | 4.1e-03 | Click! |
| chr14_17681830_17681997 | Thrb | 21017 | 0.263428 | -0.95 | 4.2e-03 | Click! |
| chr14_17685511_17685778 | Thrb | 24748 | 0.250711 | 0.94 | 4.6e-03 | Click! |
| chr14_17915471_17915812 | Thrb | 47180 | 0.161904 | -0.94 | 5.0e-03 | Click! |
Activity of the Thrb motif across conditions
Conditions sorted by the z-value of the Thrb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_126260333_126260533 | 1.81 |
Sbk1 |
SH3-binding kinase 1 |
11571 |
0.11 |
| chr14_25377112_25377271 | 1.48 |
Gm26660 |
predicted gene, 26660 |
1444 |
0.44 |
| chr19_4436418_4436609 | 1.40 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
2490 |
0.17 |
| chr19_10901185_10901365 | 1.37 |
Prpf19 |
pre-mRNA processing factor 19 |
993 |
0.37 |
| chr4_142069494_142069660 | 1.31 |
Tmem51os1 |
Tmem51 opposite strand 1 |
14395 |
0.13 |
| chr3_98288408_98289120 | 1.31 |
Gm43189 |
predicted gene 43189 |
1776 |
0.28 |
| chr16_31994081_31994256 | 1.29 |
Hmgb1-ps6 |
high mobility group box 1, pseudogene 6 |
2300 |
0.13 |
| chr19_5763306_5763457 | 1.29 |
Scyl1 |
SCY1-like 1 (S. cerevisiae) |
1605 |
0.16 |
| chr10_70099167_70099342 | 1.28 |
Ccdc6 |
coiled-coil domain containing 6 |
2133 |
0.41 |
| chr1_86600398_86600549 | 1.27 |
Cops7b |
COP9 signalosome subunit 7B |
3733 |
0.15 |
| chr12_111514450_111514666 | 1.26 |
Gm40578 |
predicted gene, 40578 |
13718 |
0.1 |
| chr4_148215189_148215340 | 1.25 |
Disp3 |
dispatched RND transporter family member 3 |
45487 |
0.08 |
| chr6_112939448_112939605 | 1.22 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
7228 |
0.15 |
| chr2_36244831_36245001 | 1.17 |
Ptgs1 |
prostaglandin-endoperoxide synthase 1 |
2441 |
0.18 |
| chr8_114982881_114983044 | 1.15 |
Gm22556 |
predicted gene, 22556 |
69951 |
0.14 |
| chr11_82015046_82015230 | 1.13 |
Gm31522 |
predicted gene, 31522 |
4829 |
0.14 |
| chr12_98573684_98573967 | 1.06 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
887 |
0.51 |
| chr2_151943059_151943241 | 1.04 |
Gm14154 |
predicted gene 14154 |
7491 |
0.14 |
| chr3_83850904_83851066 | 1.03 |
Tlr2 |
toll-like receptor 2 |
9218 |
0.2 |
| chr19_23110526_23110678 | 1.02 |
2410080I02Rik |
RIKEN cDNA 2410080I02 gene |
24298 |
0.14 |
| chr14_45315776_45315957 | 1.02 |
Ero1l |
ERO1-like (S. cerevisiae) |
2706 |
0.19 |
| chr1_191224850_191225043 | 1.01 |
D730003I15Rik |
RIKEN cDNA D730003I15 gene |
472 |
0.74 |
| chr17_13425168_13425333 | 0.99 |
Rnu6 |
U6 small nuclear RNA |
6889 |
0.12 |
| chr11_82845122_82845302 | 0.99 |
Rffl |
ring finger and FYVE like domain containing protein |
880 |
0.45 |
| chr13_56648096_56648259 | 0.98 |
Tgfbi |
transforming growth factor, beta induced |
16964 |
0.19 |
| chr2_104130173_104130336 | 0.98 |
A930018P22Rik |
RIKEN cDNA A930018P22 gene |
7485 |
0.14 |
| chr18_24652431_24652590 | 0.97 |
Mocos |
molybdenum cofactor sulfurase |
1181 |
0.43 |
| chr17_7387699_7387859 | 0.97 |
Unc93a2 |
unc-93 homolog A2 |
2315 |
0.22 |
| chr6_121890040_121890777 | 0.95 |
Mug1 |
murinoglobulin 1 |
4831 |
0.21 |
| chr5_9056138_9056296 | 0.95 |
Gm40264 |
predicted gene, 40264 |
21093 |
0.14 |
| chr5_32141078_32141248 | 0.92 |
Fosl2 |
fos-like antigen 2 |
4990 |
0.17 |
| chr10_61929391_61929572 | 0.91 |
Gm5750 |
predicted gene 5750 |
30143 |
0.16 |
| chr15_102091806_102092001 | 0.89 |
Eif4b |
eukaryotic translation initiation factor 4B |
2661 |
0.17 |
| chr3_98245800_98245972 | 0.87 |
Gm42821 |
predicted gene 42821 |
2713 |
0.21 |
| chr5_121856848_121857131 | 0.86 |
Pheta1 |
PH domain containing endocytic trafficking adaptor 1 |
7984 |
0.11 |
| chr14_26450001_26450182 | 0.86 |
Slmap |
sarcolemma associated protein |
7201 |
0.16 |
| chr3_10294464_10294624 | 0.83 |
Fabp12 |
fatty acid binding protein 12 |
6630 |
0.1 |
| chr3_36475756_36475914 | 0.82 |
Anxa5 |
annexin A5 |
59 |
0.49 |
| chr9_22420603_22420765 | 0.81 |
9530077C05Rik |
RIKEN cDNA 9530077C05 gene |
9045 |
0.11 |
| chr1_24693656_24693810 | 0.81 |
Lmbrd1 |
LMBR1 domain containing 1 |
14807 |
0.17 |
| chr7_132578212_132578403 | 0.81 |
Oat |
ornithine aminotransferase |
1909 |
0.28 |
| chr1_60824712_60825126 | 0.81 |
Gm11581 |
predicted gene 11581 |
15803 |
0.12 |
| chr15_99837964_99838156 | 0.80 |
Lima1 |
LIM domain and actin binding 1 |
17917 |
0.08 |
| chr11_109347489_109347695 | 0.78 |
1700096J18Rik |
RIKEN cDNA 1700096J18 gene |
753 |
0.57 |
| chr1_161750733_161750915 | 0.78 |
Gm5049 |
predicted gene 5049 |
4768 |
0.17 |
| chr9_57652474_57652625 | 0.78 |
Csk |
c-src tyrosine kinase |
922 |
0.44 |
| chr11_119056313_119056482 | 0.77 |
Cbx8 |
chromobox 8 |
15428 |
0.14 |
| chr3_108046974_108047188 | 0.77 |
Gstm4 |
glutathione S-transferase, mu 4 |
2187 |
0.13 |
| chr3_146846305_146846456 | 0.76 |
Ttll7 |
tubulin tyrosine ligase-like family, member 7 |
5987 |
0.18 |
| chr8_106619727_106619898 | 0.75 |
Cdh1 |
cadherin 1 |
15669 |
0.17 |
| chr15_97029707_97029895 | 0.74 |
Slc38a4 |
solute carrier family 38, member 4 |
1410 |
0.55 |
| chr11_62934808_62934982 | 0.73 |
Tvp23bos |
trans-golgi network vesicle protein 23B, opposite strand |
13303 |
0.13 |
| chr6_70814000_70814168 | 0.73 |
Rpia |
ribose 5-phosphate isomerase A |
21852 |
0.13 |
| chr9_113738969_113739195 | 0.73 |
Clasp2 |
CLIP associating protein 2 |
2391 |
0.3 |
| chr18_61374503_61374706 | 0.72 |
Gm25301 |
predicted gene, 25301 |
23209 |
0.12 |
| chr12_24630346_24630510 | 0.70 |
Gm6969 |
predicted pseudogene 6969 |
8582 |
0.16 |
| chr14_17916764_17916922 | 0.70 |
Thrb |
thyroid hormone receptor beta |
45978 |
0.16 |
| chr6_134043668_134043819 | 0.70 |
Etv6 |
ets variant 6 |
7774 |
0.2 |
| chr15_99820031_99820226 | 0.70 |
Lima1 |
LIM domain and actin binding 1 |
15 |
0.93 |
| chr13_34739256_34739451 | 0.69 |
Fam50b |
family with sequence similarity 50, member B |
259 |
0.88 |
| chr8_36202101_36202262 | 0.69 |
Gm35520 |
predicted gene, 35520 |
13809 |
0.15 |
| chr6_128498166_128498338 | 0.68 |
Pzp |
PZP, alpha-2-macroglobulin like |
3029 |
0.13 |
| chr15_75314541_75314705 | 0.68 |
Gm3454 |
predicted gene 3454 |
341 |
0.79 |
| chr4_106988955_106989270 | 0.68 |
Ssbp3 |
single-stranded DNA binding protein 3 |
22686 |
0.15 |
| chr12_85473320_85473671 | 0.67 |
Fos |
FBJ osteosarcoma oncogene |
395 |
0.83 |
| chr9_70234563_70234714 | 0.67 |
Myo1e |
myosin IE |
27270 |
0.19 |
| chr8_70657841_70658013 | 0.67 |
Pgpep1 |
pyroglutamyl-peptidase I |
489 |
0.61 |
| chr9_118958897_118959171 | 0.67 |
Ctdspl |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
29167 |
0.13 |
| chr11_60935743_60935912 | 0.66 |
Map2k3 |
mitogen-activated protein kinase kinase 3 |
3761 |
0.16 |
| chr14_63479379_63479556 | 0.66 |
Gm47074 |
predicted gene, 47074 |
4091 |
0.16 |
| chr11_71012867_71013036 | 0.66 |
Derl2 |
Der1-like domain family, member 2 |
6261 |
0.09 |
| chr14_18678485_18678659 | 0.66 |
Ube2e2 |
ubiquitin-conjugating enzyme E2E 2 |
13422 |
0.25 |
| chr4_133804499_133804683 | 0.65 |
Gm25270 |
predicted gene, 25270 |
6872 |
0.14 |
| chr7_81189147_81189298 | 0.65 |
Gm44968 |
predicted gene 44968 |
15788 |
0.13 |
| chr12_77216600_77216773 | 0.65 |
Fut8 |
fucosyltransferase 8 |
21439 |
0.19 |
| chr15_31545253_31545412 | 0.65 |
Marchf6 |
membrane associated ring-CH-type finger 6 |
14279 |
0.12 |
| chr9_107593234_107593397 | 0.64 |
Ifrd2 |
interferon-related developmental regulator 2 |
688 |
0.28 |
| chr1_131957507_131957689 | 0.64 |
Gm29103 |
predicted gene 29103 |
5356 |
0.12 |
| chr7_19744671_19744853 | 0.64 |
Nectin2 |
nectin cell adhesion molecule 2 |
4771 |
0.08 |
| chr7_38197786_38197937 | 0.63 |
D530033B14Rik |
RIKEN cDNA D530033B14 gene |
1404 |
0.32 |
| chr5_24956925_24957102 | 0.63 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
28829 |
0.14 |
| chr15_99911968_99912127 | 0.62 |
Gm20564 |
predicted gene, 20564 |
3853 |
0.1 |
| chr11_75596908_75597126 | 0.62 |
4931413K12Rik |
RIKEN cDNA 4931413K12 gene |
2246 |
0.16 |
| chr12_98860402_98860781 | 0.61 |
A930040O22Rik |
RIKEN cDNA A930040O22 gene |
28778 |
0.12 |
| chr6_122161223_122161580 | 0.61 |
Mug-ps1 |
murinoglobulin, pseudogene 1 |
14614 |
0.16 |
| chr1_186703631_186703811 | 0.61 |
Tgfb2 |
transforming growth factor, beta 2 |
612 |
0.62 |
| chr15_85808535_85808715 | 0.61 |
Cdpf1 |
cysteine rich, DPF motif domain containing 1 |
2465 |
0.2 |
| chr8_103180795_103180954 | 0.60 |
Gm45391 |
predicted gene 45391 |
35928 |
0.21 |
| chr19_58439431_58439649 | 0.60 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
14926 |
0.22 |
| chr13_43232609_43232789 | 0.60 |
Tbc1d7 |
TBC1 domain family, member 7 |
61198 |
0.1 |
| chr12_108856815_108857551 | 0.60 |
Gm22079 |
predicted gene, 22079 |
5486 |
0.1 |
| chr4_150065927_150066095 | 0.59 |
Mir34a |
microRNA 34a |
2443 |
0.16 |
| chr16_47171920_47172088 | 0.59 |
Gm18169 |
predicted gene, 18169 |
40366 |
0.21 |
| chr13_41239510_41239683 | 0.59 |
Gm32063 |
predicted gene, 32063 |
3251 |
0.15 |
| chr2_34983172_34983323 | 0.59 |
Hc |
hemolytic complement |
10089 |
0.12 |
| chr6_128483626_128483790 | 0.58 |
Pzp |
PZP, alpha-2-macroglobulin like |
4147 |
0.1 |
| chr8_126636622_126636807 | 0.58 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
42728 |
0.16 |
| chr10_80973010_80973161 | 0.57 |
Gm3828 |
predicted gene 3828 |
18375 |
0.08 |
| chr16_4886806_4886964 | 0.57 |
Mgrn1 |
mahogunin, ring finger 1 |
499 |
0.69 |
| chr12_12999123_12999274 | 0.57 |
Gm35208 |
predicted gene, 35208 |
11881 |
0.16 |
| chr18_36738903_36739066 | 0.57 |
Tmco6 |
transmembrane and coiled-coil domains 6 |
3817 |
0.09 |
| chr7_79358682_79358833 | 0.56 |
Rlbp1 |
retinaldehyde binding protein 1 |
18946 |
0.13 |
| chr19_38125391_38125548 | 0.56 |
Rbp4 |
retinol binding protein 4, plasma |
188 |
0.93 |
| chr15_66932473_66932630 | 0.55 |
Ndrg1 |
N-myc downstream regulated gene 1 |
1918 |
0.28 |
| chr11_49996483_49996658 | 0.55 |
Rnf130 |
ring finger protein 130 |
28776 |
0.14 |
| chr10_120752084_120752244 | 0.55 |
Gm10743 |
predicted gene 10743 |
3831 |
0.15 |
| chr13_98684591_98684906 | 0.55 |
Tmem171 |
transmembrane protein 171 |
10020 |
0.14 |
| chr6_88913100_88913275 | 0.54 |
Tpra1 |
transmembrane protein, adipocyte asscociated 1 |
2588 |
0.17 |
| chr13_69582483_69582651 | 0.54 |
Srd5a1 |
steroid 5 alpha-reductase 1 |
12262 |
0.12 |
| chr15_90992378_90992671 | 0.54 |
Kif21a |
kinesin family member 21A |
26122 |
0.16 |
| chr2_6595830_6596024 | 0.53 |
Celf2 |
CUGBP, Elav-like family member 2 |
3127 |
0.35 |
| chr3_101835406_101835600 | 0.53 |
Mab21l3 |
mab-21-like 3 |
720 |
0.72 |
| chr19_36692056_36692224 | 0.52 |
Hectd2os |
Hectd2, opposite strand |
2661 |
0.32 |
| chr5_139612620_139612791 | 0.52 |
Uncx |
UNC homeobox |
68807 |
0.08 |
| chr13_52153674_52153844 | 0.52 |
Gm48199 |
predicted gene, 48199 |
26652 |
0.19 |
| chr7_99641322_99641487 | 0.52 |
Gm25409 |
predicted gene, 25409 |
12414 |
0.09 |
| chr8_103538740_103538926 | 0.52 |
Gm45277 |
predicted gene 45277 |
24790 |
0.23 |
| chr9_110781917_110782081 | 0.51 |
Myl3 |
myosin, light polypeptide 3 |
13564 |
0.1 |
| chr8_12719084_12719404 | 0.51 |
Gm15348 |
predicted gene 15348 |
117 |
0.96 |
| chr10_77118451_77118858 | 0.51 |
Col18a1 |
collagen, type XVIII, alpha 1 |
4708 |
0.2 |
| chr18_75307577_75307805 | 0.51 |
2010010A06Rik |
RIKEN cDNA 2010010A06 gene |
10362 |
0.22 |
| chr2_169694792_169694990 | 0.51 |
Tshz2 |
teashirt zinc finger family member 2 |
61215 |
0.13 |
| chr2_26600848_26601015 | 0.51 |
Agpat2 |
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
2785 |
0.12 |
| chr1_71705120_71705280 | 0.51 |
Gm5829 |
predicted gene 5829 |
4855 |
0.2 |
| chr11_24789291_24789442 | 0.50 |
Gm10466 |
predicted gene 10466 |
58644 |
0.14 |
| chr2_168005469_168005791 | 0.50 |
Gm14236 |
predicted gene 14236 |
4899 |
0.17 |
| chr2_162986609_162986768 | 0.50 |
Sgk2 |
serum/glucocorticoid regulated kinase 2 |
642 |
0.6 |
| chr10_115809219_115809409 | 0.50 |
Tspan8 |
tetraspanin 8 |
7518 |
0.27 |
| chr4_152177650_152177825 | 0.50 |
Acot7 |
acyl-CoA thioesterase 7 |
397 |
0.77 |
| chr9_54560284_54560443 | 0.50 |
Cib2 |
calcium and integrin binding family member 2 |
145 |
0.95 |
| chr1_191154426_191154595 | 0.50 |
Fam71a |
family with sequence similarity 71, member A |
10307 |
0.13 |
| chr17_28290145_28290326 | 0.50 |
Ppard |
peroxisome proliferator activator receptor delta |
3011 |
0.15 |
| chr10_80928616_80928783 | 0.50 |
Gadd45b |
growth arrest and DNA-damage-inducible 45 beta |
1374 |
0.25 |
| chr6_86395136_86395301 | 0.49 |
Pcyox1 |
prenylcysteine oxidase 1 |
1408 |
0.24 |
| chr13_73723026_73723388 | 0.48 |
Slc12a7 |
solute carrier family 12, member 7 |
9887 |
0.15 |
| chr4_34966679_34966979 | 0.48 |
Gm12364 |
predicted gene 12364 |
4609 |
0.19 |
| chr16_13240471_13240636 | 0.48 |
Mrtfb |
myocardin related transcription factor B |
15928 |
0.25 |
| chr2_118554450_118554815 | 0.48 |
Bmf |
BCL2 modifying factor |
4945 |
0.18 |
| chr11_58801906_58802080 | 0.47 |
Fam183b |
family with sequence similarity 183, member B |
33 |
0.94 |
| chr1_151496001_151496317 | 0.47 |
Gm36527 |
predicted gene, 36527 |
3426 |
0.15 |
| chr6_39286782_39286965 | 0.47 |
Gm26008 |
predicted gene, 26008 |
20931 |
0.15 |
| chr13_5832208_5832483 | 0.46 |
Gm26043 |
predicted gene, 26043 |
10487 |
0.17 |
| chr6_71204222_71204852 | 0.46 |
Fabp1 |
fatty acid binding protein 1, liver |
4710 |
0.14 |
| chr16_95432244_95432403 | 0.46 |
Erg |
ETS transcription factor |
26922 |
0.22 |
| chr14_54476038_54476201 | 0.46 |
Rem2 |
rad and gem related GTP binding protein 2 |
19 |
0.95 |
| chr17_13662964_13663132 | 0.46 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
5843 |
0.16 |
| chr8_116184614_116184784 | 0.46 |
4930422C21Rik |
RIKEN cDNA 4930422C21 gene |
2670 |
0.42 |
| chr4_133028994_133029156 | 0.46 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
10418 |
0.18 |
| chr1_71648273_71648424 | 0.46 |
Apol7d |
apolipoprotein L 7d |
4489 |
0.21 |
| chr10_117082015_117082177 | 0.46 |
Cct2 |
chaperonin containing Tcp1, subunit 2 (beta) |
18282 |
0.13 |
| chr7_115825357_115825510 | 0.45 |
Sox6 |
SRY (sex determining region Y)-box 6 |
723 |
0.79 |
| chr10_115780280_115780436 | 0.45 |
Tspan8 |
tetraspanin 8 |
36474 |
0.2 |
| chr3_104670184_104670346 | 0.45 |
Gm29560 |
predicted gene 29560 |
255 |
0.84 |
| chr8_47991010_47991194 | 0.45 |
Wwc2 |
WW, C2 and coiled-coil domain containing 2 |
178 |
0.95 |
| chr10_98915744_98915895 | 0.45 |
Atp2b1 |
ATPase, Ca++ transporting, plasma membrane 1 |
347 |
0.92 |
| chr12_111956283_111956452 | 0.44 |
Gm36757 |
predicted gene, 36757 |
5850 |
0.11 |
| chr18_47219464_47219645 | 0.44 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
61641 |
0.09 |
| chr2_164563319_164563487 | 0.44 |
Wfdc2 |
WAP four-disulfide core domain 2 |
384 |
0.69 |
| chr11_109485510_109486095 | 0.44 |
Arsg |
arylsulfatase G |
196 |
0.91 |
| chr12_28579239_28579390 | 0.44 |
Allc |
allantoicase |
3201 |
0.2 |
| chr6_122249737_122249965 | 0.44 |
Mug-ps1 |
murinoglobulin, pseudogene 1 |
20791 |
0.13 |
| chr8_89364350_89364531 | 0.44 |
Gm5356 |
predicted pseudogene 5356 |
176880 |
0.03 |
| chr18_61909022_61909179 | 0.43 |
Ablim3 |
actin binding LIM protein family, member 3 |
2723 |
0.27 |
| chr19_32476509_32476692 | 0.43 |
Minpp1 |
multiple inositol polyphosphate histidine phosphatase 1 |
9169 |
0.14 |
| chr1_57989000_57989391 | 0.43 |
Sgo2a |
shugoshin 2A |
6776 |
0.19 |
| chr2_77113165_77113335 | 0.42 |
Gm44360 |
predicted gene, 44360 |
38898 |
0.16 |
| chr15_11394606_11394765 | 0.42 |
Tars |
threonyl-tRNA synthetase |
2415 |
0.37 |
| chr16_76349002_76349359 | 0.42 |
Nrip1 |
nuclear receptor interacting protein 1 |
23857 |
0.18 |
| chr3_117905348_117905499 | 0.41 |
Gm4321 |
predicted gene 4321 |
16409 |
0.2 |
| chr2_21211497_21211673 | 0.41 |
Thnsl1 |
threonine synthase-like 1 (bacterial) |
5798 |
0.17 |
| chr4_148651109_148651502 | 0.41 |
Gm572 |
predicted gene 572 |
7988 |
0.14 |
| chr6_99145207_99145385 | 0.41 |
Foxp1 |
forkhead box P1 |
17722 |
0.26 |
| chr7_126475278_126475550 | 0.41 |
Sh2b1 |
SH2B adaptor protein 1 |
10 |
0.95 |
| chr6_81944489_81944663 | 0.40 |
Gcfc2 |
GC-rich sequence DNA binding factor 2 |
1927 |
0.23 |
| chr1_127459100_127459460 | 0.40 |
Tmem163 |
transmembrane protein 163 |
41064 |
0.15 |
| chr11_63046542_63046708 | 0.40 |
Tekt3 |
tektin 3 |
15029 |
0.18 |
| chr18_60738849_60739016 | 0.40 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
5782 |
0.16 |
| chr15_25714163_25714331 | 0.40 |
Myo10 |
myosin X |
376 |
0.88 |
| chr13_37857409_37857582 | 0.40 |
Rreb1 |
ras responsive element binding protein 1 |
439 |
0.85 |
| chr1_174959965_174960140 | 0.39 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
38233 |
0.21 |
| chr9_118101918_118102083 | 0.39 |
Azi2 |
5-azacytidine induced gene 2 |
42944 |
0.13 |
| chr3_104638838_104638989 | 0.38 |
Slc16a1 |
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
245 |
0.84 |
| chr2_32248839_32249007 | 0.38 |
Pomt1 |
protein-O-mannosyltransferase 1 |
260 |
0.81 |
| chr6_72227116_72227283 | 0.38 |
Atoh8 |
atonal bHLH transcription factor 8 |
7338 |
0.17 |
| chr11_62487069_62487231 | 0.38 |
Gm12278 |
predicted gene 12278 |
4353 |
0.13 |
| chr8_116367840_116368103 | 0.38 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
10910 |
0.29 |
| chr8_70104635_70104800 | 0.38 |
Mir7066 |
microRNA 7066 |
2131 |
0.15 |
| chr15_34527692_34527846 | 0.37 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
1624 |
0.25 |
| chr5_74874877_74875112 | 0.37 |
Gm17906 |
predicted gene, 17906 |
8341 |
0.22 |
| chr1_165629214_165629365 | 0.37 |
Mpzl1 |
myelin protein zero-like 1 |
4797 |
0.13 |
| chr14_70330142_70330488 | 0.37 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
1203 |
0.35 |
| chr17_46016022_46016192 | 0.37 |
Vegfa |
vascular endothelial growth factor A |
5265 |
0.19 |
| chr1_73987938_73988111 | 0.37 |
Tns1 |
tensin 1 |
961 |
0.63 |
| chr1_52499794_52500020 | 0.37 |
Nab1 |
Ngfi-A binding protein 1 |
393 |
0.8 |
| chr6_116635450_116635635 | 0.37 |
Gm44154 |
predicted gene, 44154 |
5320 |
0.11 |
| chr5_115569868_115570025 | 0.37 |
Gcn1 |
GCN1 activator of EIF2AK4 |
4643 |
0.11 |
| chr9_83527787_83527959 | 0.37 |
Sh3bgrl2 |
SH3 domain binding glutamic acid-rich protein like 2 |
20454 |
0.14 |
| chr12_102297681_102297848 | 0.37 |
Rin3 |
Ras and Rab interactor 3 |
14123 |
0.21 |
| chr13_64362005_64362163 | 0.36 |
Gm49230 |
predicted gene, 49230 |
347 |
0.71 |
| chr10_75058843_75058998 | 0.36 |
Bcr |
BCR activator of RhoGEF and GTPase |
1672 |
0.33 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.2 | 0.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.4 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.4 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.1 | 0.5 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.3 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.1 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0006057 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.1 | 0.4 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.1 | 0.2 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.1 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.4 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.1 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.2 | GO:0052428 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.5 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.2 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.8 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.1 | GO:0060897 | neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.0 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.0 | 0.0 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.0 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.0 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0035907 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.0 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.0 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.0 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.3 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.2 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.1 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.4 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.1 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0043888 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.4 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0003933 | GTP cyclohydrolase activity(GO:0003933) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.3 | GO:0030351 | inositol-1,3,4,5,6-pentakisphosphate 3-phosphatase activity(GO:0030351) inositol-1,4,5,6-tetrakisphosphate 6-phosphatase activity(GO:0030352) inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0018595 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |