Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
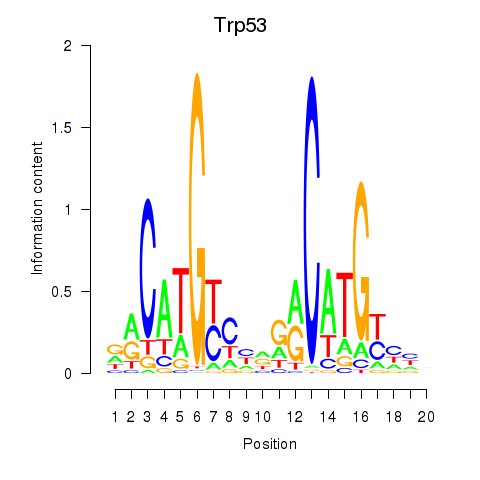
Results for Trp53
Z-value: 2.22
Transcription factors associated with Trp53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp53
|
ENSMUSG00000059552.7 | transformation related protein 53 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_69581154_69581313 | Trp53 | 828 | 0.278397 | 0.88 | 1.9e-02 | Click! |
| chr11_69581695_69581992 | Trp53 | 1438 | 0.162179 | 0.47 | 3.5e-01 | Click! |
| chr11_69580921_69581079 | Trp53 | 595 | 0.368016 | -0.42 | 4.0e-01 | Click! |
| chr11_69581394_69581562 | Trp53 | 1073 | 0.216896 | 0.21 | 6.9e-01 | Click! |
| chr11_69580228_69580399 | Trp53 | 46 | 0.493867 | 0.12 | 8.2e-01 | Click! |
Activity of the Trp53 motif across conditions
Conditions sorted by the z-value of the Trp53 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
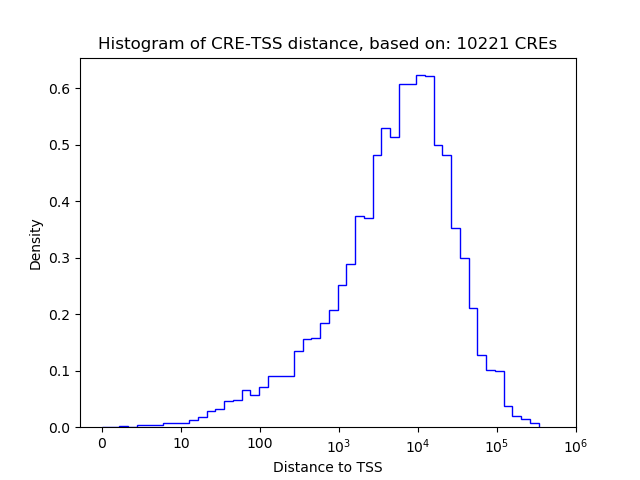
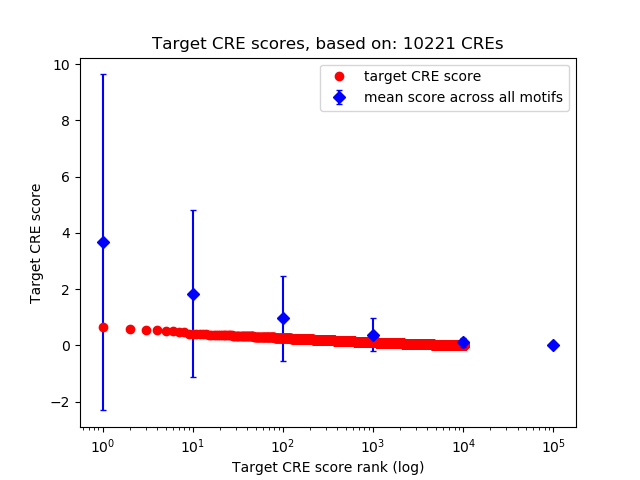
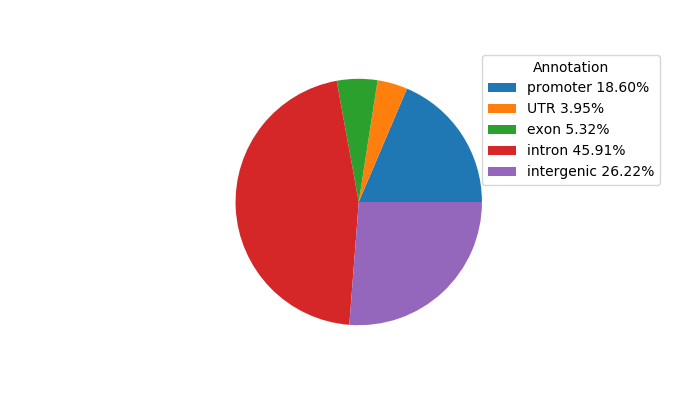
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_30525931_30526082 | 0.65 |
Tmem44 |
transmembrane protein 44 |
14903 |
0.19 |
| chr15_62039115_62039319 | 0.57 |
Pvt1 |
Pvt1 oncogene |
40 |
0.98 |
| chr13_25701872_25702071 | 0.56 |
Gm11350 |
predicted gene 11350 |
20828 |
0.27 |
| chr6_88003582_88003770 | 0.55 |
4933412L11Rik |
RIKEN cDNA 4933412L11 gene |
9497 |
0.09 |
| chr2_157471421_157471583 | 0.52 |
Src |
Rous sarcoma oncogene |
14417 |
0.16 |
| chr15_11011259_11011438 | 0.51 |
Slc45a2 |
solute carrier family 45, member 2 |
10627 |
0.16 |
| chr6_147950991_147951177 | 0.46 |
Far2 |
fatty acyl CoA reductase 2 |
96175 |
0.07 |
| chr13_93627456_93627944 | 0.46 |
Gm15622 |
predicted gene 15622 |
2318 |
0.25 |
| chr4_99227319_99227481 | 0.42 |
Atg4c |
autophagy related 4C, cysteine peptidase |
9309 |
0.19 |
| chr4_137810112_137810287 | 0.41 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
13815 |
0.19 |
| chr5_32740717_32740884 | 0.41 |
Pisd |
phosphatidylserine decarboxylase |
54 |
0.95 |
| chr6_135994801_135994964 | 0.40 |
Gm14330 |
predicted gene 14330 |
119377 |
0.06 |
| chr3_132864626_132864788 | 0.39 |
Gm29811 |
predicted gene, 29811 |
13889 |
0.14 |
| chr14_29672536_29672709 | 0.39 |
Cacna2d3 |
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
49173 |
0.13 |
| chr3_132860911_132861074 | 0.38 |
Gm29811 |
predicted gene, 29811 |
17604 |
0.14 |
| chr18_60740746_60741116 | 0.38 |
Rps14 |
ribosomal protein S14 |
6167 |
0.16 |
| chr14_69728721_69728885 | 0.38 |
Chmp7 |
charged multivesicular body protein 7 |
2072 |
0.22 |
| chr10_68269443_68270351 | 0.37 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
8824 |
0.23 |
| chr3_121356225_121356399 | 0.37 |
Gm5711 |
predicted gene 5711 |
24367 |
0.14 |
| chr17_8124666_8124826 | 0.37 |
Rnaset2a |
ribonuclease T2A |
23039 |
0.12 |
| chr10_39406363_39406707 | 0.37 |
Fyn |
Fyn proto-oncogene |
13474 |
0.19 |
| chr4_118362978_118363146 | 0.36 |
Hyi |
hydroxypyruvate isomerase (putative) |
1740 |
0.25 |
| chr3_145591903_145592058 | 0.36 |
Znhit6 |
zinc finger, HIT type 6 |
3888 |
0.24 |
| chr11_117203569_117203863 | 0.36 |
Septin9 |
septin 9 |
4055 |
0.2 |
| chr16_10539600_10539751 | 0.36 |
Dexi |
dexamethasone-induced transcript |
3379 |
0.22 |
| chr3_133765600_133766533 | 0.36 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr17_7001922_7002085 | 0.36 |
Rnaset2b |
ribonuclease T2B |
11347 |
0.13 |
| chr1_5072181_5072333 | 0.35 |
Rgs20 |
regulator of G-protein signaling 20 |
1972 |
0.26 |
| chr7_49420191_49420342 | 0.34 |
Nav2 |
neuron navigator 2 |
30494 |
0.19 |
| chr1_5071962_5072114 | 0.34 |
Rgs20 |
regulator of G-protein signaling 20 |
1753 |
0.28 |
| chr4_55014920_55015072 | 0.34 |
Zfp462 |
zinc finger protein 462 |
3516 |
0.32 |
| chr2_158629836_158630021 | 0.34 |
Actr5 |
ARP5 actin-related protein 5 |
1701 |
0.24 |
| chr11_16857621_16857825 | 0.34 |
Egfr |
epidermal growth factor receptor |
20427 |
0.18 |
| chr7_141184840_141184997 | 0.33 |
Gm22019 |
predicted gene, 22019 |
3607 |
0.09 |
| chr18_53824107_53824451 | 0.33 |
Csnk1g3 |
casein kinase 1, gamma 3 |
37843 |
0.2 |
| chr12_104901402_104901556 | 0.33 |
Gm28875 |
predicted gene 28875 |
25210 |
0.14 |
| chr2_25194509_25194660 | 0.33 |
Tor4a |
torsin family 4, member A |
2175 |
0.1 |
| chr11_103047839_103047990 | 0.33 |
Nmt1 |
N-myristoyltransferase 1 |
19329 |
0.1 |
| chr1_93210442_93210625 | 0.33 |
Sned1 |
sushi, nidogen and EGF-like domains 1 |
25308 |
0.11 |
| chr8_11373743_11373894 | 0.32 |
Col4a2 |
collagen, type IV, alpha 2 |
31090 |
0.13 |
| chr6_50363496_50363647 | 0.32 |
Osbpl3 |
oxysterol binding protein-like 3 |
6644 |
0.27 |
| chr19_3881058_3881209 | 0.32 |
Chka |
choline kinase alpha |
5728 |
0.09 |
| chr3_123128727_123129043 | 0.32 |
Synpo2 |
synaptopodin 2 |
11833 |
0.14 |
| chr2_151977784_151977939 | 0.32 |
Fam110a |
family with sequence similarity 110, member A |
2358 |
0.21 |
| chr6_5045977_5046153 | 0.32 |
Ppp1r9a |
protein phosphatase 1, regulatory subunit 9A |
64594 |
0.1 |
| chr14_64198612_64198817 | 0.32 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
82400 |
0.07 |
| chr4_135780360_135780511 | 0.32 |
Myom3 |
myomesin family, member 3 |
17755 |
0.12 |
| chr19_24268067_24268282 | 0.31 |
Fxn |
frataxin |
9997 |
0.17 |
| chr5_34346535_34346723 | 0.31 |
Rnf4 |
ring finger protein 4 |
4487 |
0.15 |
| chr13_92660201_92660368 | 0.31 |
Serinc5 |
serine incorporator 5 |
45498 |
0.14 |
| chr9_102639370_102639523 | 0.31 |
Anapc13 |
anaphase promoting complex subunit 13 |
10934 |
0.13 |
| chr4_130129683_130130093 | 0.31 |
Hcrtr1 |
hypocretin (orexin) receptor 1 |
7740 |
0.15 |
| chr12_25817244_25817410 | 0.31 |
Gm9321 |
predicted gene 9321 |
3214 |
0.34 |
| chr12_7990141_7990303 | 0.31 |
Apob |
apolipoprotein B |
12443 |
0.21 |
| chr7_80653537_80653707 | 0.31 |
Gm15880 |
predicted gene 15880 |
17605 |
0.15 |
| chr2_71719599_71719934 | 0.31 |
Platr26 |
pluripotency associated transcript 26 |
349 |
0.83 |
| chr1_136142734_136142885 | 0.31 |
Kif21b |
kinesin family member 21B |
11355 |
0.11 |
| chr11_120005928_120006087 | 0.30 |
Aatk |
apoptosis-associated tyrosine kinase |
6401 |
0.11 |
| chr16_77434703_77434862 | 0.30 |
9430053O09Rik |
RIKEN cDNA 9430053O09 gene |
12962 |
0.11 |
| chr4_108754553_108754711 | 0.30 |
Gm12741 |
predicted gene 12741 |
22890 |
0.12 |
| chr2_84626485_84626636 | 0.30 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
11545 |
0.1 |
| chr11_99177164_99177321 | 0.30 |
Ccr7 |
chemokine (C-C motif) receptor 7 |
22165 |
0.13 |
| chr14_63822463_63822616 | 0.30 |
Xkr6 |
X-linked Kx blood group related 6 |
27520 |
0.17 |
| chr6_126806165_126806322 | 0.30 |
Gm29788 |
predicted gene, 29788 |
1873 |
0.27 |
| chr5_119054472_119054644 | 0.30 |
1700081H04Rik |
RIKEN cDNA 1700081H04 gene |
53676 |
0.13 |
| chr16_10436240_10436428 | 0.30 |
Tvp23a |
trans-golgi network vesicle protein 23A |
7073 |
0.16 |
| chr2_103028308_103028465 | 0.30 |
Pdhx |
pyruvate dehydrogenase complex, component X |
44949 |
0.13 |
| chr1_127400359_127400512 | 0.30 |
Gm38301 |
predicted gene, 38301 |
6341 |
0.21 |
| chr2_84937644_84937820 | 0.30 |
Slc43a3 |
solute carrier family 43, member 3 |
842 |
0.5 |
| chr5_114806082_114806287 | 0.29 |
Ankrd13a |
ankyrin repeat domain 13a |
6613 |
0.09 |
| chr4_148427588_148427746 | 0.29 |
Gm23303 |
predicted gene, 23303 |
15818 |
0.13 |
| chr5_103737634_103737904 | 0.29 |
Aff1 |
AF4/FMR2 family, member 1 |
16393 |
0.19 |
| chr5_140438868_140439039 | 0.29 |
Eif3b |
eukaryotic translation initiation factor 3, subunit B |
989 |
0.42 |
| chr15_64195944_64196287 | 0.29 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
12376 |
0.24 |
| chr3_138291566_138291979 | 0.28 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
14121 |
0.11 |
| chr13_64314711_64314862 | 0.28 |
Prxl2c |
peroxiredoxin like 2C |
2076 |
0.17 |
| chr18_56490748_56490899 | 0.28 |
Gramd3 |
GRAM domain containing 3 |
21556 |
0.14 |
| chr7_145056434_145056606 | 0.28 |
Gm45181 |
predicted gene 45181 |
106476 |
0.05 |
| chr9_44665366_44665550 | 0.28 |
Gm3953 |
predicted gene 3953 |
742 |
0.4 |
| chr5_114596263_114596414 | 0.28 |
Fam222a |
family with sequence similarity 222, member A |
28321 |
0.12 |
| chr2_93432778_93432949 | 0.28 |
Mir7001 |
microRNA 7001 |
10856 |
0.15 |
| chr14_63145558_63145861 | 0.28 |
Ctsb |
cathepsin B |
10191 |
0.13 |
| chr5_140831353_140831504 | 0.27 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
997 |
0.6 |
| chr5_123364346_123364497 | 0.27 |
5830487J09Rik |
RIKEN cDNA 5830487J09 gene |
2051 |
0.15 |
| chr8_11305954_11306113 | 0.27 |
Col4a1 |
collagen, type IV, alpha 1 |
6656 |
0.17 |
| chr17_33539247_33539403 | 0.27 |
Adamts10 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
1606 |
0.31 |
| chr6_133960296_133960447 | 0.27 |
Gm8956 |
predicted gene 8956 |
27962 |
0.17 |
| chr13_29769741_29769892 | 0.27 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
85615 |
0.1 |
| chr6_121069331_121069498 | 0.27 |
Gm4651 |
predicted gene 4651 |
4818 |
0.19 |
| chr3_152754407_152754729 | 0.27 |
Pigk |
phosphatidylinositol glycan anchor biosynthesis, class K |
8813 |
0.21 |
| chr7_140383247_140383438 | 0.27 |
Gm6376 |
predicted gene 6376 |
3002 |
0.12 |
| chr4_118149267_118149447 | 0.27 |
Kdm4a |
lysine (K)-specific demethylase 4A |
2598 |
0.22 |
| chr15_74784791_74785147 | 0.27 |
Gm17189 |
predicted gene 17189 |
904 |
0.33 |
| chr5_74660839_74660990 | 0.26 |
Lnx1 |
ligand of numb-protein X 1 |
16715 |
0.17 |
| chr13_56324320_56324471 | 0.26 |
Gm17878 |
predicted gene, 17878 |
4061 |
0.17 |
| chr3_100485827_100486059 | 0.26 |
Tent5c |
terminal nucleotidyltransferase 5C |
3251 |
0.18 |
| chr13_93626823_93626974 | 0.26 |
Gm15622 |
predicted gene 15622 |
1516 |
0.35 |
| chr8_111820161_111820326 | 0.26 |
Cfdp1 |
craniofacial development protein 1 |
34048 |
0.13 |
| chr10_127632738_127633305 | 0.26 |
Gm48815 |
predicted gene, 48815 |
3366 |
0.11 |
| chr17_66041118_66041288 | 0.26 |
Ankrd12 |
ankyrin repeat domain 12 |
35261 |
0.1 |
| chr16_30057093_30057267 | 0.26 |
Hes1 |
hes family bHLH transcription factor 1 |
7204 |
0.16 |
| chr12_79347763_79347934 | 0.26 |
Rad51b |
RAD51 paralog B |
20495 |
0.2 |
| chr6_119643983_119644181 | 0.26 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
21977 |
0.22 |
| chr1_64791430_64791588 | 0.26 |
Plekhm3 |
pleckstrin homology domain containing, family M, member 3 |
45255 |
0.11 |
| chr10_41796051_41796202 | 0.26 |
Cep57l1 |
centrosomal protein 57-like 1 |
13481 |
0.15 |
| chr7_98312602_98312888 | 0.26 |
Acer3 |
alkaline ceramidase 3 |
3207 |
0.25 |
| chr5_33574701_33574864 | 0.26 |
Fam53a |
family with sequence similarity 53, member A |
54130 |
0.08 |
| chr7_142375895_142376054 | 0.25 |
Ifitm10 |
interferon induced transmembrane protein 10 |
2221 |
0.16 |
| chr4_33244314_33244465 | 0.25 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
3186 |
0.21 |
| chr13_34281709_34281879 | 0.25 |
Gm47086 |
predicted gene, 47086 |
37014 |
0.14 |
| chr12_103362325_103362476 | 0.25 |
Gm45777 |
predicted gene 45777 |
6372 |
0.11 |
| chr5_27411249_27411405 | 0.25 |
Speer4b |
spermatogenesis associated glutamate (E)-rich protein 4B |
90035 |
0.09 |
| chr18_76000614_76000773 | 0.25 |
1700003O11Rik |
RIKEN cDNA 1700003O11 gene |
14984 |
0.23 |
| chr12_86799109_86799540 | 0.25 |
Gm10095 |
predicted gene 10095 |
47143 |
0.11 |
| chr14_30934111_30934339 | 0.25 |
Itih1 |
inter-alpha trypsin inhibitor, heavy chain 1 |
9052 |
0.1 |
| chr13_70967895_70968046 | 0.25 |
8030423J24Rik |
RIKEN cDNA 8030423J24 gene |
85022 |
0.08 |
| chr19_12457466_12457644 | 0.25 |
Mpeg1 |
macrophage expressed gene 1 |
3224 |
0.15 |
| chr1_136704194_136704370 | 0.25 |
Gm24086 |
predicted gene, 24086 |
6908 |
0.14 |
| chr8_104831387_104831616 | 0.25 |
Ces2b |
carboxyesterase 2B |
71 |
0.94 |
| chr9_74884431_74884582 | 0.24 |
Onecut1 |
one cut domain, family member 1 |
18022 |
0.15 |
| chr19_60941165_60941317 | 0.24 |
Gm9529 |
predicted gene 9529 |
26011 |
0.14 |
| chr6_38186452_38186609 | 0.24 |
D630045J12Rik |
RIKEN cDNA D630045J12 gene |
7892 |
0.2 |
| chr6_91525048_91525199 | 0.24 |
Gm45216 |
predicted gene 45216 |
2714 |
0.17 |
| chr14_65682096_65682315 | 0.24 |
Scara5 |
scavenger receptor class A, member 5 |
5728 |
0.25 |
| chr7_66371115_66371303 | 0.24 |
Gm23957 |
predicted gene, 23957 |
728 |
0.56 |
| chr1_58891620_58891795 | 0.24 |
Gm28802 |
predicted gene 28802 |
1085 |
0.48 |
| chr6_77240615_77240802 | 0.24 |
Lrrtm1 |
leucine rich repeat transmembrane neuronal 1 |
1981 |
0.43 |
| chr3_151840491_151840642 | 0.24 |
Ptgfr |
prostaglandin F receptor |
2936 |
0.22 |
| chr2_116932327_116932524 | 0.24 |
D330050G23Rik |
RIKEN cDNA D330050G23 gene |
32231 |
0.13 |
| chr6_54767552_54767839 | 0.24 |
Znrf2 |
zinc and ring finger 2 |
49221 |
0.12 |
| chr6_6538473_6538631 | 0.24 |
Sem1 |
SEM1, 26S proteasome complex subunit |
40106 |
0.14 |
| chr12_78842565_78842734 | 0.24 |
Atp6v1d |
ATPase, H+ transporting, lysosomal V1 subunit D |
2742 |
0.23 |
| chr8_83104070_83104221 | 0.24 |
Gm45449 |
predicted gene 45449 |
17188 |
0.2 |
| chrX_42018134_42018310 | 0.24 |
Xiap |
X-linked inhibitor of apoptosis |
41457 |
0.14 |
| chr9_114716436_114716587 | 0.24 |
Cmtm6 |
CKLF-like MARVEL transmembrane domain containing 6 |
14605 |
0.15 |
| chr13_63665698_63665976 | 0.24 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr7_45574063_45574214 | 0.24 |
Bcat2 |
branched chain aminotransferase 2, mitochondrial |
962 |
0.22 |
| chr6_86145033_86145200 | 0.24 |
Gm19596 |
predicted gene, 19596 |
2036 |
0.27 |
| chr11_20370399_20370788 | 0.24 |
Gm12033 |
predicted gene 12033 |
34580 |
0.15 |
| chr2_173531317_173531484 | 0.24 |
1700021F07Rik |
RIKEN cDNA 1700021F07 gene |
8814 |
0.21 |
| chr15_25623084_25623317 | 0.23 |
Myo10 |
myosin X |
651 |
0.7 |
| chr12_80947275_80947910 | 0.23 |
Srsf5 |
serine and arginine-rich splicing factor 5 |
1216 |
0.34 |
| chr9_34735078_34735235 | 0.23 |
Kirrel3os |
kirre like nephrin family adhesion molecule 3, opposite strand |
194353 |
0.03 |
| chr18_46714530_46714763 | 0.23 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
13383 |
0.13 |
| chr4_139614424_139614599 | 0.23 |
Aldh4a1 |
aldehyde dehydrogenase 4 family, member A1 |
8355 |
0.14 |
| chr11_58969062_58969229 | 0.23 |
Trim17 |
tripartite motif-containing 17 |
5343 |
0.07 |
| chr14_70395125_70395415 | 0.23 |
Gm22725 |
predicted gene, 22725 |
27010 |
0.09 |
| chr16_87512452_87512619 | 0.23 |
Gm24891 |
predicted gene, 24891 |
15630 |
0.12 |
| chr9_65299789_65300058 | 0.23 |
Gm16218 |
predicted gene 16218 |
2004 |
0.18 |
| chr2_72741921_72742104 | 0.23 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
8841 |
0.22 |
| chr6_35333770_35333951 | 0.23 |
1700065J11Rik |
RIKEN cDNA 1700065J11 gene |
3014 |
0.23 |
| chr5_145983671_145983967 | 0.23 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
7824 |
0.13 |
| chr2_13235859_13236020 | 0.23 |
Gm37160 |
predicted gene, 37160 |
4688 |
0.22 |
| chr9_53344725_53344910 | 0.23 |
Exph5 |
exophilin 5 |
3628 |
0.22 |
| chr1_60899201_60899544 | 0.23 |
Rpl18-ps1 |
ribosomal protein L18, pseudogene 1 |
1526 |
0.31 |
| chr11_31771118_31771290 | 0.23 |
D630024D03Rik |
RIKEN cDNA D630024D03 gene |
53269 |
0.12 |
| chr12_32829679_32829842 | 0.23 |
Nampt |
nicotinamide phosphoribosyltransferase |
8959 |
0.2 |
| chr11_81370503_81370687 | 0.23 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
14418 |
0.29 |
| chr16_23272256_23272412 | 0.23 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
2705 |
0.23 |
| chr12_111454567_111454858 | 0.23 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
4193 |
0.14 |
| chr18_80468518_80468718 | 0.23 |
Gm50209 |
predicted gene, 50209 |
239 |
0.87 |
| chr17_27587363_27587662 | 0.23 |
Nudt3 |
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
4193 |
0.1 |
| chr2_163685840_163686012 | 0.23 |
Gm16316 |
predicted gene 16316 |
6353 |
0.15 |
| chr6_86003630_86003791 | 0.22 |
4930553P18Rik |
RIKEN cDNA 4930553P18 gene |
670 |
0.54 |
| chr4_128840187_128840338 | 0.22 |
Gm12968 |
predicted gene 12968 |
5935 |
0.17 |
| chr12_44248581_44248746 | 0.22 |
Gm47096 |
predicted gene, 47096 |
20455 |
0.12 |
| chr2_132123048_132123340 | 0.22 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
11746 |
0.16 |
| chr5_118010944_118011095 | 0.22 |
Gm42545 |
predicted gene 42545 |
16351 |
0.13 |
| chr3_65935257_65935428 | 0.22 |
Gm37037 |
predicted gene, 37037 |
1434 |
0.3 |
| chr1_172296958_172297109 | 0.22 |
Atp1a2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
1031 |
0.37 |
| chr9_70761117_70761279 | 0.22 |
Adam10 |
a disintegrin and metallopeptidase domain 10 |
14855 |
0.2 |
| chr16_86824945_86825096 | 0.22 |
Gm32624 |
predicted gene, 32624 |
8146 |
0.19 |
| chr3_65533694_65533867 | 0.22 |
4931440P22Rik |
RIKEN cDNA 4931440P22 gene |
4401 |
0.15 |
| chr17_55511801_55511952 | 0.22 |
St6gal2 |
beta galactoside alpha 2,6 sialyltransferase 2 |
65632 |
0.11 |
| chr5_137828792_137828976 | 0.22 |
Pilra |
paired immunoglobin-like type 2 receptor alpha |
2626 |
0.12 |
| chr2_129526434_129526601 | 0.22 |
F830045P16Rik |
RIKEN cDNA F830045P16 gene |
10085 |
0.16 |
| chr4_154705761_154705932 | 0.22 |
Actrt2 |
actin-related protein T2 |
37979 |
0.12 |
| chr12_3344324_3344475 | 0.22 |
Gm26520 |
predicted gene, 26520 |
12754 |
0.13 |
| chr2_104131753_104132101 | 0.22 |
A930018P22Rik |
RIKEN cDNA A930018P22 gene |
9158 |
0.14 |
| chr4_118152144_118152311 | 0.22 |
Kdm4a |
lysine (K)-specific demethylase 4A |
5468 |
0.16 |
| chr14_70575919_70576089 | 0.22 |
Nudt18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
1633 |
0.23 |
| chr10_121485281_121485446 | 0.22 |
Gm40787 |
predicted gene, 40787 |
3623 |
0.14 |
| chr3_92440282_92440451 | 0.22 |
Sprr1b |
small proline-rich protein 1B |
1577 |
0.18 |
| chr3_86135244_86135409 | 0.22 |
Snord73a |
small nucleolar RNA, C/D box U73A |
3532 |
0.12 |
| chr6_141879515_141879720 | 0.22 |
Gm30784 |
predicted gene, 30784 |
13259 |
0.2 |
| chr5_123025352_123025521 | 0.22 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
10098 |
0.08 |
| chr11_6389224_6389393 | 0.22 |
Zmiz2 |
zinc finger, MIZ-type containing 2 |
56 |
0.95 |
| chr10_84054746_84054912 | 0.22 |
Gm37908 |
predicted gene, 37908 |
6929 |
0.2 |
| chr11_102263318_102263476 | 0.21 |
Hrob |
homologous recombination factor with OB-fold |
1433 |
0.24 |
| chr19_10940905_10941056 | 0.21 |
Ptgdr2 |
prostaglandin D2 receptor 2 |
3820 |
0.12 |
| chr14_51008118_51008288 | 0.21 |
Rnase10 |
ribonuclease, RNase A family, 10 (non-active) |
274 |
0.8 |
| chr7_19876833_19877030 | 0.21 |
Gm44659 |
predicted gene 44659 |
5493 |
0.08 |
| chr18_21372400_21372792 | 0.21 |
Gm22886 |
predicted gene, 22886 |
8335 |
0.2 |
| chr8_109667901_109668069 | 0.21 |
Ist1 |
increased sodium tolerance 1 homolog (yeast) |
9609 |
0.15 |
| chr10_71252490_71252774 | 0.21 |
Ube2d1 |
ubiquitin-conjugating enzyme E2D 1 |
4256 |
0.15 |
| chr17_56687946_56688097 | 0.21 |
Ranbp3 |
RAN binding protein 3 |
8714 |
0.11 |
| chr14_25793517_25793703 | 0.21 |
Gm27185 |
predicted gene 27185 |
16271 |
0.13 |
| chr13_47215938_47216142 | 0.21 |
Rnf144b |
ring finger protein 144B |
22215 |
0.17 |
| chr3_10129292_10129733 | 0.21 |
Gm37308 |
predicted gene, 37308 |
39557 |
0.1 |
| chr5_113152964_113153205 | 0.21 |
Gm42161 |
predicted gene, 42161 |
7756 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.3 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.2 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.0 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0061620 | glycolytic process through glucose-6-phosphate(GO:0061620) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0002585 | positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.0 | 0.0 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.0 | 0.0 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) regulation of cortisol biosynthetic process(GO:2000064) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) |
| 0.0 | 0.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0043328 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:0034114 | regulation of heterotypic cell-cell adhesion(GO:0034114) |
| 0.0 | 0.0 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.0 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.0 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.0 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0052687 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0044606 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0043888 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |