Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
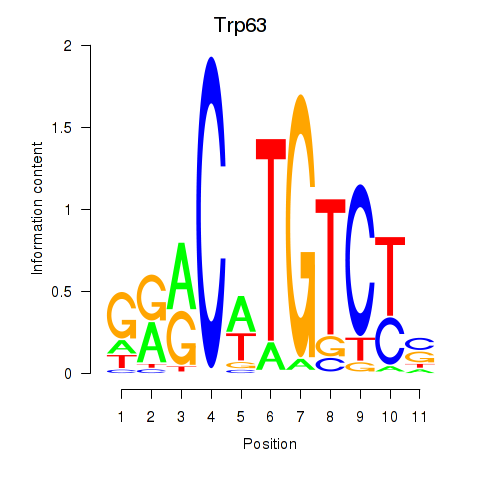
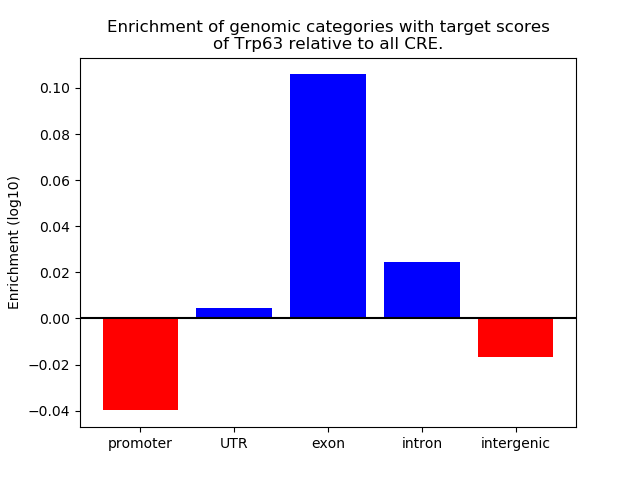
Results for Trp63
Z-value: 1.39
Transcription factors associated with Trp63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp63
|
ENSMUSG00000022510.8 | transformation related protein 63 |
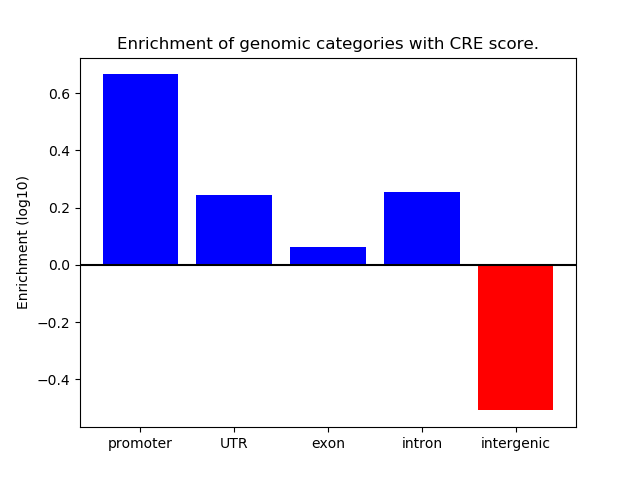
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_25875334_25875496 | Trp63 | 73499 | 0.102754 | 0.95 | 3.6e-03 | Click! |
| chr16_25876965_25877159 | Trp63 | 75146 | 0.099620 | 0.89 | 1.8e-02 | Click! |
| chr16_25805292_25805462 | Trp63 | 3461 | 0.331388 | 0.88 | 2.0e-02 | Click! |
| chr16_25876261_25876542 | Trp63 | 74485 | 0.100867 | 0.88 | 2.0e-02 | Click! |
| chr16_25804778_25805017 | Trp63 | 2981 | 0.351663 | 0.77 | 7.1e-02 | Click! |
Activity of the Trp63 motif across conditions
Conditions sorted by the z-value of the Trp63 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
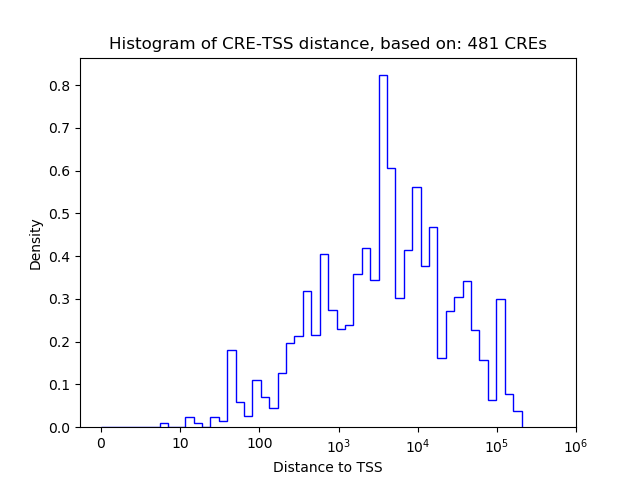
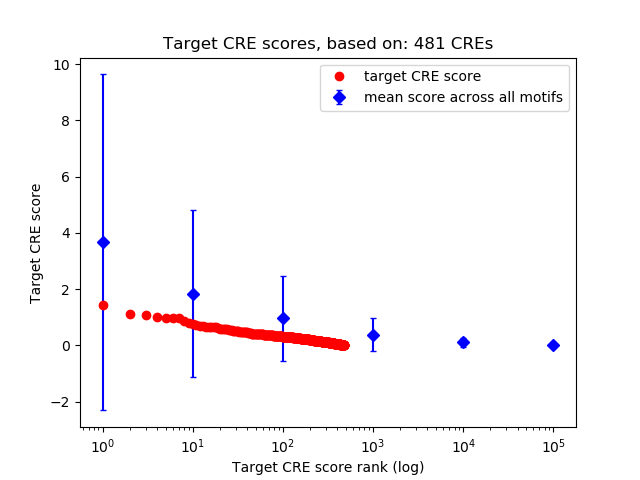
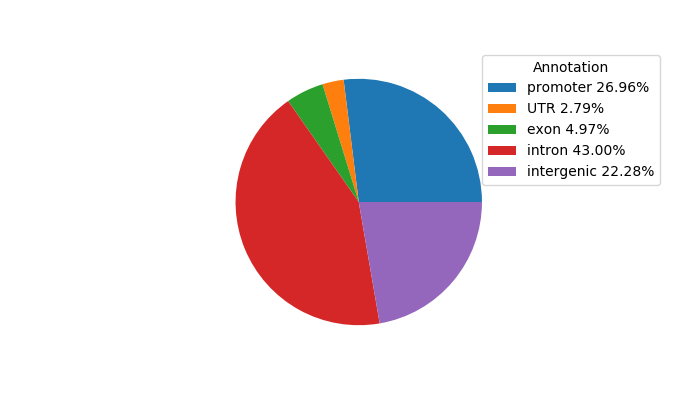
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_104083020_104083260 | 1.44 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr5_125512652_125512844 | 1.11 |
Aacs |
acetoacetyl-CoA synthetase |
49 |
0.97 |
| chr1_88122933_88123203 | 1.08 |
Gm15372 |
predicted gene 15372 |
3708 |
0.08 |
| chr7_98355230_98355493 | 1.01 |
Tsku |
tsukushi, small leucine rich proteoglycan |
4718 |
0.19 |
| chr14_21181534_21181685 | 0.97 |
Adk |
adenosine kinase |
105457 |
0.06 |
| chr19_40184875_40185171 | 0.96 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
2263 |
0.24 |
| chr15_59015531_59015876 | 0.96 |
Mtss1 |
MTSS I-BAR domain containing 1 |
24893 |
0.16 |
| chr8_70476439_70476616 | 0.87 |
Klhl26 |
kelch-like 26 |
378 |
0.72 |
| chr7_99201987_99202161 | 0.79 |
Gm45012 |
predicted gene 45012 |
292 |
0.86 |
| chr7_98355533_98355727 | 0.77 |
Tsku |
tsukushi, small leucine rich proteoglycan |
4449 |
0.2 |
| chr15_3398924_3399075 | 0.71 |
Ghr |
growth hormone receptor |
72645 |
0.11 |
| chr5_140112771_140112922 | 0.67 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
2620 |
0.23 |
| chr11_70501874_70502034 | 0.67 |
Tm4sf5 |
transmembrane 4 superfamily member 5 |
3290 |
0.09 |
| chr18_9842900_9843072 | 0.67 |
Gm23637 |
predicted gene, 23637 |
15867 |
0.13 |
| chr16_42911354_42911708 | 0.65 |
Gm19522 |
predicted gene, 19522 |
534 |
0.77 |
| chr12_75566070_75566284 | 0.65 |
Gm47689 |
predicted gene, 47689 |
13688 |
0.18 |
| chr4_134902978_134903198 | 0.64 |
Tmem50a |
transmembrane protein 50A |
11927 |
0.14 |
| chr11_11845438_11845699 | 0.63 |
Ddc |
dopa decarboxylase |
9288 |
0.18 |
| chr9_48722520_48722671 | 0.63 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
113350 |
0.05 |
| chr16_18071409_18071560 | 0.59 |
Dgcr6 |
DiGeorge syndrome critical region gene 6 |
2004 |
0.23 |
| chr11_5530853_5531004 | 0.58 |
Xbp1 |
X-box binding protein 1 |
9005 |
0.13 |
| chr19_39282659_39282846 | 0.57 |
Cyp2c29 |
cytochrome P450, family 2, subfamily c, polypeptide 29 |
4322 |
0.25 |
| chr17_89646332_89646522 | 0.57 |
Gm50061 |
predicted gene, 50061 |
106099 |
0.08 |
| chr6_35873017_35873173 | 0.57 |
Gm43442 |
predicted gene 43442 |
53616 |
0.16 |
| chr8_104789386_104789560 | 0.56 |
Gm45782 |
predicted gene 45782 |
3645 |
0.12 |
| chr5_120481198_120481714 | 0.53 |
Gm15690 |
predicted gene 15690 |
553 |
0.54 |
| chr19_46056737_46057343 | 0.53 |
Pprc1 |
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
176 |
0.92 |
| chr11_16781411_16781603 | 0.52 |
Egfr |
epidermal growth factor receptor |
29277 |
0.16 |
| chr11_120816054_120816209 | 0.50 |
Fasn |
fatty acid synthase |
676 |
0.49 |
| chr5_92585412_92585563 | 0.50 |
Fam47e |
family with sequence similarity 47, member E |
13931 |
0.15 |
| chr11_16841273_16841607 | 0.49 |
Egfros |
epidermal growth factor receptor, opposite strand |
10738 |
0.21 |
| chr13_41202938_41203089 | 0.49 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
17149 |
0.11 |
| chr10_87913477_87913628 | 0.49 |
Igf1os |
insulin-like growth factor 1, opposite strand |
50171 |
0.11 |
| chr7_19018429_19018664 | 0.49 |
Foxa3 |
forkhead box A3 |
4992 |
0.08 |
| chr3_138226457_138226622 | 0.48 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
1512 |
0.29 |
| chr10_31595125_31595561 | 0.47 |
Rnf217 |
ring finger protein 217 |
13841 |
0.16 |
| chr6_95710009_95710160 | 0.47 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
8716 |
0.32 |
| chr9_53292248_53292436 | 0.47 |
Exph5 |
exophilin 5 |
9328 |
0.19 |
| chr6_100096782_100096939 | 0.47 |
2010109P13Rik |
RIKEN cDNA 2010109P13 gene |
46954 |
0.15 |
| chr14_64690169_64690374 | 0.46 |
Kif13b |
kinesin family member 13B |
37669 |
0.15 |
| chr11_68851235_68851391 | 0.46 |
Ndel1 |
nudE neurodevelopment protein 1 like 1 |
1728 |
0.27 |
| chr6_85868321_85868637 | 0.44 |
Nat8f2 |
N-acetyltransferase 8 (GCN5-related) family member 2 |
679 |
0.48 |
| chr11_5916279_5916587 | 0.43 |
Gm11967 |
predicted gene 11967 |
1118 |
0.3 |
| chr15_89077796_89078211 | 0.43 |
Trabd |
TraB domain containing |
1879 |
0.16 |
| chr2_134550332_134550521 | 0.42 |
Hao1 |
hydroxyacid oxidase 1, liver |
3881 |
0.35 |
| chr6_149098639_149098790 | 0.42 |
Dennd5b |
DENN/MADD domain containing 5B |
2824 |
0.16 |
| chr2_24901182_24901348 | 0.42 |
Gm37139 |
predicted gene, 37139 |
7399 |
0.1 |
| chr6_90568851_90569300 | 0.41 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
1369 |
0.35 |
| chr12_81167975_81168146 | 0.41 |
Mir3067 |
microRNA 3067 |
1909 |
0.38 |
| chr4_45852505_45852662 | 0.41 |
Stra6l |
STRA6-like |
3586 |
0.2 |
| chr6_82647878_82648248 | 0.40 |
Pole4 |
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
4455 |
0.23 |
| chr1_59157668_59157869 | 0.40 |
Mpp4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
893 |
0.43 |
| chr1_169165587_169165766 | 0.40 |
Mir6354 |
microRNA 6354 |
146587 |
0.04 |
| chr1_171168499_171168673 | 0.40 |
Mpz |
myelin protein zero |
8883 |
0.09 |
| chr8_109998736_109999105 | 0.40 |
Tat |
tyrosine aminotransferase |
8414 |
0.12 |
| chr2_158570467_158570618 | 0.39 |
Arhgap40 |
Rho GTPase activating protein 40 |
28805 |
0.1 |
| chr1_71723212_71723548 | 0.39 |
Gm5256 |
predicted gene 5256 |
8456 |
0.19 |
| chr4_95625117_95625268 | 0.39 |
Fggy |
FGGY carbohydrate kinase domain containing |
1636 |
0.46 |
| chr14_21256205_21256412 | 0.39 |
Adk |
adenosine kinase |
61817 |
0.13 |
| chr9_61809239_61809390 | 0.39 |
Gm19208 |
predicted gene, 19208 |
42390 |
0.15 |
| chr7_136457949_136458342 | 0.38 |
Gm36849 |
predicted gene, 36849 |
104781 |
0.07 |
| chr14_55147433_55147865 | 0.38 |
Gm23596 |
predicted gene, 23596 |
13959 |
0.09 |
| chr2_26593869_26594069 | 0.38 |
Egfl7 |
EGF-like domain 7 |
1822 |
0.16 |
| chr1_151131178_151131360 | 0.38 |
Gm8941 |
predicted gene 8941 |
5238 |
0.13 |
| chr9_57059997_57060182 | 0.37 |
Ptpn9 |
protein tyrosine phosphatase, non-receptor type 9 |
651 |
0.62 |
| chrX_60504109_60504260 | 0.37 |
Gm7073 |
predicted gene 7073 |
35065 |
0.14 |
| chr7_121652771_121652941 | 0.37 |
Usp31 |
ubiquitin specific peptidase 31 |
13959 |
0.18 |
| chr11_16589151_16589356 | 0.37 |
Gm12663 |
predicted gene 12663 |
46813 |
0.12 |
| chrX_161717217_161717797 | 0.37 |
Rai2 |
retinoic acid induced 2 |
120 |
0.98 |
| chr3_95882847_95883034 | 0.36 |
Ciart |
circadian associated repressor of transcription |
689 |
0.44 |
| chr1_136625692_136626183 | 0.36 |
Zfp281 |
zinc finger protein 281 |
1036 |
0.34 |
| chr15_10632975_10633126 | 0.36 |
Gm10389 |
predicted gene 10389 |
32713 |
0.14 |
| chr5_125478134_125478423 | 0.36 |
Gm27551 |
predicted gene, 27551 |
1099 |
0.36 |
| chr9_40804280_40804486 | 0.36 |
Hspa8 |
heat shock protein 8 |
101 |
0.66 |
| chr5_114155622_114155805 | 0.35 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
9178 |
0.12 |
| chr2_160340207_160340432 | 0.35 |
Gm826 |
predicted gene 826 |
6157 |
0.26 |
| chr6_149344317_149344633 | 0.35 |
Gm15782 |
predicted gene 15782 |
9351 |
0.14 |
| chr9_115307686_115307849 | 0.35 |
Stt3b |
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) |
2654 |
0.24 |
| chr8_85500511_85501282 | 0.34 |
Gpt2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
8287 |
0.17 |
| chr13_111895157_111895352 | 0.34 |
Gm9025 |
predicted gene 9025 |
11077 |
0.15 |
| chr18_74301601_74301911 | 0.34 |
Gm14328 |
predicted gene 14328 |
2175 |
0.26 |
| chr2_59160190_59160538 | 0.34 |
Ccdc148 |
coiled-coil domain containing 148 |
170 |
0.8 |
| chr10_37139807_37140183 | 0.34 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
81 |
0.95 |
| chr12_70198492_70198710 | 0.33 |
Pygl |
liver glycogen phosphorylase |
760 |
0.59 |
| chr6_58733664_58733815 | 0.33 |
Gm43899 |
predicted gene, 43899 |
48198 |
0.13 |
| chr6_108426569_108426787 | 0.33 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
8801 |
0.22 |
| chr11_6528187_6528435 | 0.33 |
Snora9 |
small nucleolar RNA, H/ACA box 9 |
146 |
0.77 |
| chr15_68363065_68363728 | 0.33 |
Gm20732 |
predicted gene 20732 |
220 |
0.9 |
| chr11_79878516_79878681 | 0.33 |
9130204K15Rik |
RIKEN cDNA 9130204K15 gene |
6204 |
0.15 |
| chrX_60400421_60400586 | 0.33 |
Atp11c |
ATPase, class VI, type 11C |
3478 |
0.24 |
| chr2_31514792_31514964 | 0.32 |
Ass1 |
argininosuccinate synthetase 1 |
3612 |
0.21 |
| chr13_73767240_73767553 | 0.32 |
Slc12a7 |
solute carrier family 12, member 7 |
3657 |
0.21 |
| chr12_16584004_16584353 | 0.32 |
Lpin1 |
lipin 1 |
5542 |
0.26 |
| chr4_148590620_148590972 | 0.32 |
Srm |
spermidine synthase |
707 |
0.49 |
| chr6_118478873_118479106 | 0.32 |
Zfp9 |
zinc finger protein 9 |
331 |
0.85 |
| chr4_10943152_10943325 | 0.32 |
Rps11-ps3 |
ribosomal protein S11, pseudogene 3 |
3370 |
0.23 |
| chr4_53073780_53074212 | 0.32 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
33070 |
0.15 |
| chr11_115418711_115418869 | 0.32 |
Atp5h |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
49 |
0.92 |
| chr1_194360305_194360456 | 0.31 |
4930503O07Rik |
RIKEN cDNA 4930503O07 gene |
137621 |
0.05 |
| chr17_5232385_5232541 | 0.31 |
Gm15599 |
predicted gene 15599 |
120353 |
0.05 |
| chr5_99247519_99247696 | 0.31 |
Rasgef1b |
RasGEF domain family, member 1B |
4367 |
0.26 |
| chr8_24575832_24575983 | 0.31 |
Ido2 |
indoleamine 2,3-dioxygenase 2 |
426 |
0.82 |
| chr19_55110942_55111169 | 0.30 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
7964 |
0.2 |
| chr12_74320624_74320791 | 0.30 |
Gm34552 |
predicted gene, 34552 |
3545 |
0.18 |
| chr4_14805319_14805470 | 0.30 |
Lrrc69 |
leucine rich repeat containing 69 |
9334 |
0.19 |
| chr9_69226678_69226839 | 0.30 |
Rora |
RAR-related orphan receptor alpha |
31217 |
0.22 |
| chr12_87393286_87393480 | 0.30 |
Gm48873 |
predicted gene, 48873 |
1061 |
0.3 |
| chr4_119550388_119550568 | 0.30 |
Gm12959 |
predicted gene 12959 |
3580 |
0.12 |
| chr7_141092673_141092858 | 0.30 |
Sigirr |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
985 |
0.3 |
| chr12_80324041_80324208 | 0.30 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
52568 |
0.08 |
| chr17_34853353_34853526 | 0.30 |
Nelfe |
negative elongation factor complex member E, Rdbp |
497 |
0.46 |
| chr5_112276946_112277327 | 0.30 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
429 |
0.74 |
| chr8_127111900_127112059 | 0.30 |
Pard3 |
par-3 family cell polarity regulator |
38739 |
0.16 |
| chr5_122455478_122455753 | 0.30 |
Anapc7 |
anaphase promoting complex subunit 7 |
14240 |
0.09 |
| chr12_8811934_8812098 | 0.29 |
Sdc1 |
syndecan 1 |
40213 |
0.13 |
| chr1_130664035_130664240 | 0.29 |
C4bp |
complement component 4 binding protein |
2505 |
0.18 |
| chr1_191225182_191225346 | 0.29 |
D730003I15Rik |
RIKEN cDNA D730003I15 gene |
790 |
0.54 |
| chr12_28804906_28805226 | 0.29 |
Gm48905 |
predicted gene, 48905 |
5072 |
0.18 |
| chr13_75844352_75844786 | 0.29 |
Glrx |
glutaredoxin |
4493 |
0.17 |
| chr17_45957936_45958127 | 0.28 |
Gm49805 |
predicted gene, 49805 |
3828 |
0.22 |
| chr15_78947569_78947720 | 0.28 |
Triobp |
TRIO and F-actin binding protein |
80 |
0.93 |
| chr5_124233026_124233422 | 0.28 |
Pitpnm2os1 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 1 |
3499 |
0.14 |
| chr15_99042750_99042942 | 0.28 |
Prph |
peripherin |
12328 |
0.08 |
| chr14_118038826_118039199 | 0.28 |
Dct |
dopachrome tautomerase |
4117 |
0.23 |
| chr2_91930123_91930286 | 0.28 |
Mdk |
midkine |
1484 |
0.25 |
| chr12_77056757_77057146 | 0.28 |
Gm35189 |
predicted gene, 35189 |
14750 |
0.21 |
| chr10_80433536_80433845 | 0.28 |
Tcf3 |
transcription factor 3 |
43 |
0.94 |
| chr1_163150756_163150961 | 0.28 |
Gm22434 |
predicted gene, 22434 |
32631 |
0.15 |
| chr8_111693865_111694016 | 0.28 |
Ctrb1 |
chymotrypsinogen B1 |
2930 |
0.21 |
| chr12_76369660_76370048 | 0.28 |
Zbtb25 |
zinc finger and BTB domain containing 25 |
252 |
0.62 |
| chr14_51932437_51932626 | 0.28 |
Gm49076 |
predicted gene, 49076 |
2871 |
0.12 |
| chr5_138776683_138776881 | 0.27 |
Fam20c |
family with sequence similarity 20, member C |
9718 |
0.19 |
| chr1_76911315_76911480 | 0.27 |
Gm816 |
predicted gene 816 |
116395 |
0.07 |
| chr17_35814760_35815402 | 0.27 |
Ier3 |
immediate early response 3 |
6603 |
0.07 |
| chr19_55548698_55548991 | 0.27 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
167830 |
0.03 |
| chr9_30855695_30855884 | 0.27 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
53868 |
0.12 |
| chr8_93283453_93283604 | 0.27 |
Ces1f |
carboxylesterase 1F |
3781 |
0.19 |
| chr8_80813537_80813711 | 0.26 |
Gab1 |
growth factor receptor bound protein 2-associated protein 1 |
17260 |
0.18 |
| chr10_59323965_59324136 | 0.26 |
P4ha1 |
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
682 |
0.72 |
| chr5_120849731_120849899 | 0.26 |
Oas1f |
2'-5' oligoadenylate synthetase 1F |
1443 |
0.23 |
| chr17_35254463_35254614 | 0.26 |
Mir8094 |
microRNA 8094 |
3249 |
0.07 |
| chr16_21787535_21788055 | 0.25 |
Ehhadh |
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
12 |
0.96 |
| chr17_25861995_25862180 | 0.25 |
Wdr90 |
WD repeat domain 90 |
586 |
0.4 |
| chr1_131980100_131980306 | 0.25 |
Slc45a3 |
solute carrier family 45, member 3 |
1239 |
0.34 |
| chr15_85658037_85658248 | 0.25 |
Lncppara |
long noncoding RNA near Ppara |
4526 |
0.17 |
| chr17_46073702_46073982 | 0.25 |
Gm36200 |
predicted gene, 36200 |
10424 |
0.13 |
| chr2_155674493_155674644 | 0.25 |
Trpc4ap |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
1517 |
0.26 |
| chr1_91465824_91465979 | 0.25 |
Gm28381 |
predicted gene 28381 |
4197 |
0.13 |
| chr16_62846272_62846452 | 0.25 |
Arl13b |
ADP-ribosylation factor-like 13B |
631 |
0.69 |
| chr16_30283830_30283981 | 0.24 |
Lrrc15 |
leucine rich repeat containing 15 |
649 |
0.64 |
| chr15_88861401_88861700 | 0.24 |
Pim3 |
proviral integration site 3 |
636 |
0.63 |
| chr16_18428559_18428717 | 0.24 |
Txnrd2 |
thioredoxin reductase 2 |
59 |
0.94 |
| chr6_52606167_52606352 | 0.24 |
Gm44434 |
predicted gene, 44434 |
3014 |
0.21 |
| chr15_80679418_80679782 | 0.24 |
Fam83f |
family with sequence similarity 83, member F |
7753 |
0.13 |
| chr14_17900415_17900617 | 0.24 |
Thrb |
thyroid hormone receptor beta |
62305 |
0.13 |
| chr14_27328803_27328977 | 0.24 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
7176 |
0.23 |
| chr1_160692487_160692638 | 0.24 |
Gm37328 |
predicted gene, 37328 |
34119 |
0.09 |
| chr17_43113552_43113703 | 0.24 |
E130008D07Rik |
RIKEN cDNA E130008D07 gene |
44569 |
0.18 |
| chr3_88536664_88536842 | 0.24 |
Mir1905 |
microRNA 1905 |
371 |
0.68 |
| chr8_109982750_109983074 | 0.24 |
Tat |
tyrosine aminotransferase |
7525 |
0.12 |
| chr13_60730621_60730876 | 0.23 |
Dapk1 |
death associated protein kinase 1 |
23248 |
0.14 |
| chr5_138948024_138948178 | 0.23 |
Pdgfa |
platelet derived growth factor, alpha |
46180 |
0.12 |
| chr6_145855716_145856207 | 0.23 |
Gm43909 |
predicted gene, 43909 |
7336 |
0.17 |
| chr7_101301283_101301902 | 0.23 |
Atg16l2 |
autophagy related 16-like 2 (S. cerevisiae) |
435 |
0.73 |
| chr1_86497756_86497986 | 0.23 |
Rpl30-ps6 |
ribosomal protein L30, pseudogene 6 |
24212 |
0.11 |
| chr7_128460851_128461002 | 0.23 |
Tial1 |
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
283 |
0.79 |
| chr5_140119388_140119599 | 0.23 |
Mad1l1 |
MAD1 mitotic arrest deficient 1-like 1 |
4027 |
0.19 |
| chr4_115411935_115412583 | 0.23 |
Cyp4a12b |
cytochrome P450, family 4, subfamily a, polypeptide 12B |
635 |
0.62 |
| chr6_146470035_146470223 | 0.23 |
Gm15720 |
predicted gene 15720 |
6522 |
0.21 |
| chr4_102430791_102430956 | 0.23 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
826 |
0.77 |
| chr7_51972606_51972763 | 0.23 |
Gas2 |
growth arrest specific 2 |
29000 |
0.14 |
| chr1_175607741_175607962 | 0.23 |
Fh1 |
fumarate hydratase 1 |
1204 |
0.45 |
| chr14_41109398_41109729 | 0.22 |
Mat1a |
methionine adenosyltransferase I, alpha |
4182 |
0.14 |
| chr4_45467831_45468030 | 0.22 |
Shb |
src homology 2 domain-containing transforming protein B |
14872 |
0.15 |
| chr13_99092586_99092968 | 0.22 |
2310020H05Rik |
RIKEN cDNA 2310020H05 gene |
4856 |
0.18 |
| chr2_30981618_30981782 | 0.22 |
BC005624 |
cDNA sequence BC005624 |
241 |
0.74 |
| chr4_131989644_131989821 | 0.22 |
Epb41 |
erythrocyte membrane protein band 4.1 |
1554 |
0.27 |
| chr19_14598748_14598937 | 0.22 |
Tle4 |
transducin-like enhancer of split 4 |
791 |
0.75 |
| chr13_119617412_119617581 | 0.22 |
Gm48265 |
predicted gene, 48265 |
4024 |
0.15 |
| chr7_49399643_49399820 | 0.22 |
Nav2 |
neuron navigator 2 |
35010 |
0.18 |
| chr2_127507226_127507383 | 0.21 |
AL731831.1 |
prominin 2 (Prom2), pseudogene |
11991 |
0.13 |
| chr11_119109577_119109728 | 0.21 |
Gm11754 |
predicted gene 11754 |
1773 |
0.27 |
| chr12_70719191_70719367 | 0.21 |
Gm32369 |
predicted gene, 32369 |
37429 |
0.13 |
| chr8_106838845_106839041 | 0.21 |
Has3 |
hyaluronan synthase 3 |
31299 |
0.13 |
| chr11_69686428_69686593 | 0.21 |
Tnfsf13 |
tumor necrosis factor (ligand) superfamily, member 13 |
726 |
0.29 |
| chr7_118698775_118698967 | 0.21 |
Gde1 |
glycerophosphodiester phosphodiesterase 1 |
6509 |
0.13 |
| chr8_105811176_105811604 | 0.21 |
Ranbp10 |
RAN binding protein 10 |
15815 |
0.09 |
| chr6_119351793_119352010 | 0.21 |
Cacna2d4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
19339 |
0.17 |
| chr2_136952741_136952943 | 0.21 |
Slx4ip |
SLX4 interacting protein |
47299 |
0.13 |
| chr15_3559431_3559670 | 0.21 |
Ghr |
growth hormone receptor |
22292 |
0.23 |
| chr3_152478706_152478877 | 0.21 |
Ak5 |
adenylate kinase 5 |
276 |
0.91 |
| chr13_44733163_44733323 | 0.21 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
797 |
0.73 |
| chr2_102694561_102694718 | 0.21 |
Slc1a2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
11717 |
0.23 |
| chr11_98863094_98863383 | 0.21 |
Wipf2 |
WAS/WASL interacting protein family, member 2 |
400 |
0.74 |
| chr12_76264161_76264635 | 0.20 |
Mthfd1 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
8918 |
0.12 |
| chr9_59542656_59542807 | 0.20 |
Tmem202 |
transmembrane protein 202 |
2884 |
0.18 |
| chr19_58372705_58372871 | 0.20 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
81678 |
0.09 |
| chr8_69959520_69959671 | 0.20 |
Gatad2a |
GATA zinc finger domain containing 2A |
1191 |
0.37 |
| chr7_136894564_136895115 | 0.20 |
Mgmt |
O-6-methylguanine-DNA methyltransferase |
225 |
0.95 |
| chr15_97247773_97248330 | 0.19 |
Amigo2 |
adhesion molecule with Ig like domain 2 |
764 |
0.48 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.1 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.2 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 1.1 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0009078 | alanine metabolic process(GO:0006522) pyruvate family amino acid metabolic process(GO:0009078) |
| 0.0 | 0.1 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.0 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0043915 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0090409 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |