Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
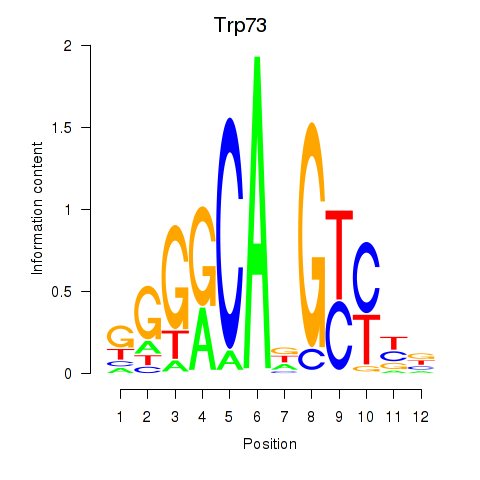
Results for Trp73
Z-value: 1.52
Transcription factors associated with Trp73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp73
|
ENSMUSG00000029026.10 | transformation related protein 73 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_154081057_154081217 | Trp73 | 4185 | 0.129702 | 0.90 | 1.6e-02 | Click! |
| chr4_154105367_154105533 | Trp73 | 8277 | 0.114828 | -0.78 | 6.7e-02 | Click! |
| chr4_154118930_154119081 | Trp73 | 2400 | 0.180872 | -0.15 | 7.8e-01 | Click! |
Activity of the Trp73 motif across conditions
Conditions sorted by the z-value of the Trp73 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
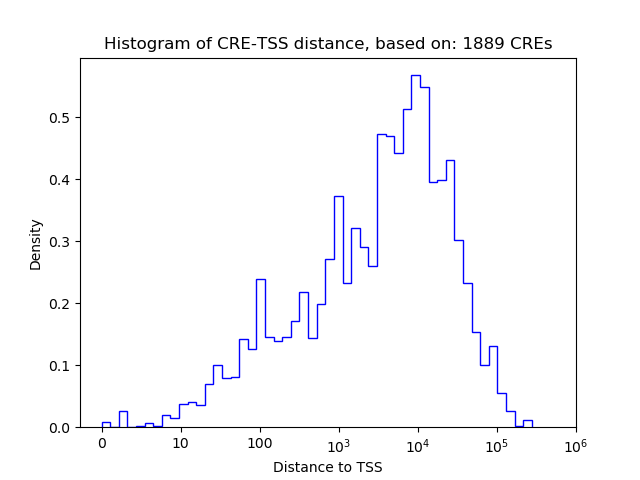
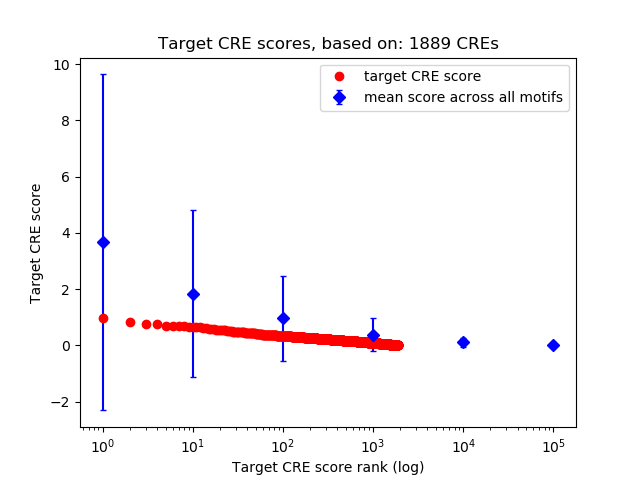
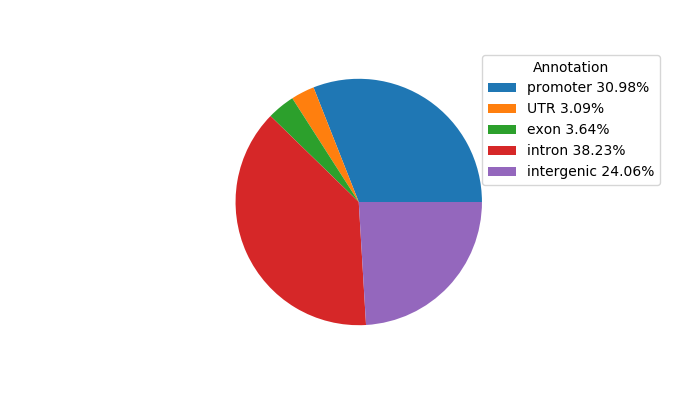
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_72243451_72243602 | 0.96 |
1700051A21Rik |
RIKEN cDNA 1700051A21 gene |
22895 |
0.09 |
| chr14_66672460_66672626 | 0.82 |
Adra1a |
adrenergic receptor, alpha 1a |
36987 |
0.17 |
| chr5_134155757_134155938 | 0.77 |
Rcc1l |
reculator of chromosome condensation 1 like |
12413 |
0.13 |
| chr11_107703521_107703697 | 0.74 |
Cacng1 |
calcium channel, voltage-dependent, gamma subunit 1 |
12913 |
0.16 |
| chr7_145069187_145069341 | 0.70 |
Gm45181 |
predicted gene 45181 |
93732 |
0.07 |
| chr17_84183727_84183878 | 0.69 |
Gm36279 |
predicted gene, 36279 |
1954 |
0.27 |
| chr6_4749062_4749219 | 0.68 |
Peg10 |
paternally expressed 10 |
1734 |
0.27 |
| chr13_28930434_28930607 | 0.68 |
Sox4 |
SRY (sex determining region Y)-box 4 |
23193 |
0.17 |
| chr4_57320936_57321111 | 0.66 |
Ptpn3 |
protein tyrosine phosphatase, non-receptor type 3 |
19186 |
0.15 |
| chr15_31568768_31568949 | 0.66 |
Cmbl |
carboxymethylenebutenolidase-like (Pseudomonas) |
60 |
0.96 |
| chr8_46385009_46385281 | 0.64 |
Gm45253 |
predicted gene 45253 |
743 |
0.6 |
| chr9_66802090_66802293 | 0.63 |
BC050972 |
cDNA sequence BC050972 |
368 |
0.78 |
| chr10_41884316_41884467 | 0.62 |
Sesn1 |
sestrin 1 |
3048 |
0.28 |
| chrX_12249219_12249376 | 0.62 |
Gm14521 |
predicted gene 14521 |
69230 |
0.11 |
| chr12_75746027_75746188 | 0.58 |
Sgpp1 |
sphingosine-1-phosphate phosphatase 1 |
10378 |
0.23 |
| chr9_123237941_123238104 | 0.58 |
Cdcp1 |
CUB domain containing protein 1 |
21984 |
0.13 |
| chr5_116390234_116390471 | 0.56 |
4930562A09Rik |
RIKEN cDNA 4930562A09 gene |
2448 |
0.2 |
| chr4_148455809_148455972 | 0.56 |
Mtor |
mechanistic target of rapamycin kinase |
7265 |
0.14 |
| chr9_43673281_43673432 | 0.56 |
Gm5364 |
predicted gene 5364 |
9697 |
0.16 |
| chr4_59331623_59331836 | 0.55 |
Susd1 |
sushi domain containing 1 |
1293 |
0.48 |
| chr15_40124618_40124784 | 0.55 |
9330182O14Rik |
RIKEN cDNA 9330182O14 gene |
9664 |
0.19 |
| chr7_46830137_46830394 | 0.54 |
Gm45308 |
predicted gene 45308 |
2199 |
0.15 |
| chr17_8343757_8343973 | 0.53 |
Prr18 |
proline rich 18 |
3126 |
0.15 |
| chr11_76893564_76893941 | 0.52 |
Tmigd1 |
transmembrane and immunoglobulin domain containing 1 |
8407 |
0.18 |
| chr3_122513095_122513259 | 0.51 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
955 |
0.47 |
| chr8_88555776_88556143 | 0.50 |
Gm45496 |
predicted gene 45496 |
4074 |
0.24 |
| chr7_132729545_132729708 | 0.49 |
Fam53b |
family with sequence similarity 53, member B |
47290 |
0.12 |
| chr8_126839083_126839255 | 0.49 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
64 |
0.98 |
| chr15_54963168_54963544 | 0.49 |
Gm26684 |
predicted gene, 26684 |
10408 |
0.15 |
| chr9_63201496_63201967 | 0.48 |
Skor1 |
SKI family transcriptional corepressor 1 |
52770 |
0.12 |
| chr1_134232873_134233068 | 0.47 |
Adora1 |
adenosine A1 receptor |
1608 |
0.3 |
| chr3_105705277_105705434 | 0.47 |
Inka2 |
inka box actin regulator 2 |
103 |
0.95 |
| chr3_83040525_83040907 | 0.47 |
Fgb |
fibrinogen beta chain |
9147 |
0.14 |
| chr10_20673746_20674078 | 0.47 |
Gm17230 |
predicted gene 17230 |
48277 |
0.14 |
| chr12_8778168_8778353 | 0.46 |
Sdc1 |
syndecan 1 |
6457 |
0.19 |
| chr2_23071515_23071743 | 0.46 |
Acbd5 |
acyl-Coenzyme A binding domain containing 5 |
2178 |
0.29 |
| chr12_103437726_103437893 | 0.46 |
Ifi27 |
interferon, alpha-inducible protein 27 |
182 |
0.89 |
| chr7_129790460_129790637 | 0.45 |
Gm44778 |
predicted gene 44778 |
42498 |
0.18 |
| chr2_173152634_173153061 | 0.45 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
201 |
0.93 |
| chr10_128961226_128961428 | 0.45 |
Mettl7b |
methyltransferase like 7B |
339 |
0.74 |
| chr6_145477400_145477576 | 0.44 |
Gm25373 |
predicted gene, 25373 |
13299 |
0.17 |
| chr19_23110526_23110678 | 0.44 |
2410080I02Rik |
RIKEN cDNA 2410080I02 gene |
24298 |
0.14 |
| chr1_78860116_78860328 | 0.44 |
Gm29187 |
predicted gene 29187 |
38964 |
0.15 |
| chr12_94190897_94191174 | 0.44 |
Gm18749 |
predicted gene, 18749 |
131334 |
0.05 |
| chr7_19785264_19785423 | 0.44 |
Cblc |
Casitas B-lineage lymphoma c |
97 |
0.91 |
| chr18_68011811_68011962 | 0.43 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
78629 |
0.09 |
| chr8_120856918_120857076 | 0.43 |
Gm26878 |
predicted gene, 26878 |
23209 |
0.19 |
| chr9_88697034_88697219 | 0.42 |
Gm37226 |
predicted gene, 37226 |
10940 |
0.1 |
| chr17_27557519_27557704 | 0.42 |
Hmga1 |
high mobility group AT-hook 1 |
916 |
0.28 |
| chr10_127962820_127963065 | 0.42 |
Gm47949 |
predicted gene, 47949 |
1386 |
0.24 |
| chr10_17551883_17552066 | 0.41 |
Gm47770 |
predicted gene, 47770 |
25060 |
0.17 |
| chr13_28629708_28629873 | 0.41 |
Mir6368 |
microRNA 6368 |
81083 |
0.09 |
| chr2_32607757_32607919 | 0.41 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
233 |
0.82 |
| chr9_65461146_65461312 | 0.41 |
Spg21 |
SPG21, maspardin |
276 |
0.85 |
| chr6_88627459_88627634 | 0.41 |
Kbtbd12 |
kelch repeat and BTB (POZ) domain containing 12 |
98 |
0.97 |
| chr1_93479013_93479164 | 0.40 |
Septin2 |
septin 2 |
20 |
0.79 |
| chr2_168003768_168003979 | 0.40 |
Gm14236 |
predicted gene 14236 |
6656 |
0.16 |
| chr6_113828811_113828998 | 0.39 |
Gm44167 |
predicted gene, 44167 |
29220 |
0.13 |
| chr7_87246448_87246638 | 0.38 |
Nox4 |
NADPH oxidase 4 |
106 |
0.97 |
| chr4_130112167_130112332 | 0.38 |
Pef1 |
penta-EF hand domain containing 1 |
4693 |
0.16 |
| chr15_27634069_27634229 | 0.38 |
Gm46506 |
predicted gene, 46506 |
2362 |
0.24 |
| chr5_120611405_120611556 | 0.38 |
Rita1 |
RBPJ interacting and tubulin associated 1 |
157 |
0.87 |
| chr3_22078075_22078419 | 0.38 |
Tbl1xr1 |
transducin (beta)-like 1X-linked receptor 1 |
963 |
0.47 |
| chr8_119400211_119400396 | 0.38 |
Mlycd |
malonyl-CoA decarboxylase |
5405 |
0.17 |
| chr16_95769487_95769687 | 0.37 |
Gm37259 |
predicted gene, 37259 |
9751 |
0.17 |
| chr12_3894495_3894825 | 0.37 |
Dnmt3a |
DNA methyltransferase 3A |
164 |
0.95 |
| chr3_52897779_52897957 | 0.37 |
Gm20750 |
predicted gene, 20750 |
29798 |
0.16 |
| chr7_112170278_112170643 | 0.37 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
11403 |
0.25 |
| chr6_50986659_50986810 | 0.37 |
Gm44402 |
predicted gene, 44402 |
19466 |
0.22 |
| chr17_29431823_29432064 | 0.37 |
Gm36199 |
predicted gene, 36199 |
892 |
0.45 |
| chr16_23063931_23064082 | 0.36 |
Kng1 |
kininogen 1 |
5613 |
0.08 |
| chr14_69732071_69732222 | 0.36 |
Chmp7 |
charged multivesicular body protein 7 |
388 |
0.79 |
| chr9_61479695_61479879 | 0.36 |
Gm47240 |
predicted gene, 47240 |
6801 |
0.2 |
| chr16_21866250_21866424 | 0.36 |
Gm26744 |
predicted gene, 26744 |
20561 |
0.11 |
| chr9_43465193_43465354 | 0.36 |
Gm28215 |
predicted gene 28215 |
4149 |
0.22 |
| chr12_86888572_86888744 | 0.36 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
3860 |
0.22 |
| chr5_142440070_142440526 | 0.36 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
23646 |
0.17 |
| chr14_39195795_39195969 | 0.35 |
Gm20642 |
predicted gene 20642 |
24189 |
0.28 |
| chr17_43326768_43326935 | 0.35 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
33600 |
0.19 |
| chr7_132245773_132246102 | 0.35 |
Chst15 |
carbohydrate sulfotransferase 15 |
32688 |
0.14 |
| chr15_79327440_79327599 | 0.35 |
Pla2g6 |
phospholipase A2, group VI |
445 |
0.7 |
| chr1_36316895_36317108 | 0.35 |
Arid5a |
AT rich interactive domain 5A (MRF1-like) |
26 |
0.97 |
| chr5_149014913_149015064 | 0.35 |
Gm15411 |
predicted gene 15411 |
796 |
0.43 |
| chr5_7955475_7955629 | 0.35 |
Gm30835 |
predicted gene, 30835 |
4794 |
0.18 |
| chr15_75675616_75675767 | 0.35 |
Top1mt |
DNA topoisomerase 1, mitochondrial |
3109 |
0.15 |
| chr16_31994081_31994256 | 0.35 |
Hmgb1-ps6 |
high mobility group box 1, pseudogene 6 |
2300 |
0.13 |
| chr8_10879580_10879754 | 0.35 |
B930025P03Rik |
RIKEN cDNA B930025P03 gene |
2768 |
0.15 |
| chr5_122730871_122731058 | 0.35 |
P2rx4 |
purinergic receptor P2X, ligand-gated ion channel 4 |
3300 |
0.16 |
| chr10_93282645_93282796 | 0.34 |
Elk3 |
ELK3, member of ETS oncogene family |
28091 |
0.14 |
| chr5_63936466_63936784 | 0.34 |
Rell1 |
RELT-like 1 |
5711 |
0.18 |
| chr10_93513920_93514254 | 0.34 |
Hal |
histidine ammonia lyase |
12962 |
0.13 |
| chr13_52048885_52049060 | 0.34 |
Gm37872 |
predicted gene, 37872 |
27045 |
0.17 |
| chr4_155564029_155564190 | 0.34 |
Nadk |
NAD kinase |
37 |
0.96 |
| chr5_65107589_65107740 | 0.34 |
Klhl5 |
kelch-like 5 |
125 |
0.96 |
| chr15_57979415_57979570 | 0.34 |
Fam83a |
family with sequence similarity 83, member A |
5927 |
0.18 |
| chr2_125263901_125264052 | 0.34 |
A530010F05Rik |
RIKEN cDNA A530010F05 gene |
10588 |
0.16 |
| chr16_23997987_23998171 | 0.33 |
Bcl6 |
B cell leukemia/lymphoma 6 |
9227 |
0.16 |
| chr4_83270585_83270742 | 0.33 |
Ttc39b |
tetratricopeptide repeat domain 39B |
4624 |
0.22 |
| chr19_23071512_23071878 | 0.33 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
4158 |
0.22 |
| chr11_3255972_3256130 | 0.33 |
Drg1 |
developmentally regulated GTP binding protein 1 |
3192 |
0.16 |
| chr7_45782387_45782600 | 0.33 |
Lmtk3 |
lemur tyrosine kinase 3 |
1245 |
0.21 |
| chr8_111624424_111624606 | 0.33 |
Ldhd |
lactate dehydrogenase D |
4072 |
0.18 |
| chr6_145469440_145469652 | 0.33 |
Gm25373 |
predicted gene, 25373 |
5357 |
0.18 |
| chr2_163736041_163736204 | 0.33 |
Ada |
adenosine deaminase |
5256 |
0.18 |
| chr11_69601181_69601332 | 0.33 |
Atp1b2 |
ATPase, Na+/K+ transporting, beta 2 polypeptide |
1485 |
0.15 |
| chr2_29295626_29296066 | 0.33 |
6530402F18Rik |
RIKEN cDNA 6530402F18 gene |
42840 |
0.12 |
| chrX_159256995_159257386 | 0.33 |
Rps6ka3 |
ribosomal protein S6 kinase polypeptide 3 |
1064 |
0.63 |
| chr8_34455567_34455746 | 0.32 |
Gm45349 |
predicted gene 45349 |
17339 |
0.17 |
| chr8_113141217_113141373 | 0.32 |
Gm10280 |
predicted gene 10280 |
74033 |
0.12 |
| chr4_144977157_144977352 | 0.32 |
Vps13d |
vacuolar protein sorting 13D |
6707 |
0.21 |
| chr7_130855841_130856000 | 0.32 |
Plekha1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
9836 |
0.16 |
| chr3_84324115_84324266 | 0.32 |
Trim2 |
tripartite motif-containing 2 |
17313 |
0.23 |
| chr4_138972772_138972945 | 0.32 |
Tmco4 |
transmembrane and coiled-coil domains 4 |
30 |
0.97 |
| chr11_69416113_69416276 | 0.32 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
2519 |
0.13 |
| chr15_88704398_88704558 | 0.32 |
Brd1 |
bromodomain containing 1 |
3614 |
0.26 |
| chr4_117978300_117978463 | 0.32 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
3345 |
0.19 |
| chr1_136955166_136955491 | 0.32 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
1698 |
0.42 |
| chr2_33177813_33178235 | 0.32 |
Gm13528 |
predicted gene 13528 |
122 |
0.95 |
| chr10_37837777_37837948 | 0.32 |
Gm24710 |
predicted gene, 24710 |
93818 |
0.09 |
| chr11_97437884_97438051 | 0.32 |
Arhgap23 |
Rho GTPase activating protein 23 |
1682 |
0.32 |
| chr14_73445827_73446221 | 0.32 |
Gm4266 |
predicted gene 4266 |
47069 |
0.11 |
| chr3_105895379_105895692 | 0.32 |
Adora3 |
adenosine A3 receptor |
8886 |
0.12 |
| chr10_5823911_5824075 | 0.31 |
Mtrf1l |
mitochondrial translational release factor 1-like |
83 |
0.98 |
| chr9_108095047_108095215 | 0.31 |
Apeh |
acylpeptide hydrolase |
525 |
0.53 |
| chr5_134554086_134554240 | 0.31 |
Clip2 |
CAP-GLY domain containing linker protein 2 |
1729 |
0.19 |
| chr3_10298177_10298449 | 0.31 |
Fabp12 |
fatty acid binding protein 12 |
2861 |
0.14 |
| chr13_59009996_59010157 | 0.31 |
Gm34245 |
predicted gene, 34245 |
68220 |
0.09 |
| chr2_167831188_167831379 | 0.31 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
2363 |
0.25 |
| chr9_67617399_67617691 | 0.31 |
Tln2 |
talin 2 |
57842 |
0.12 |
| chr12_85170010_85170261 | 0.31 |
Pgf |
placental growth factor |
240 |
0.87 |
| chr9_13890286_13890446 | 0.31 |
Gm7588 |
predicted gene 7588 |
21660 |
0.15 |
| chr15_78340778_78340947 | 0.31 |
Mir7676-2 |
microRNA 7676-2 |
7737 |
0.12 |
| chr8_115718761_115718923 | 0.31 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
11048 |
0.23 |
| chr18_65024180_65024941 | 0.31 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
836 |
0.66 |
| chr15_12209919_12210084 | 0.31 |
Mtmr12 |
myotubularin related protein 12 |
4883 |
0.13 |
| chr8_41035719_41035932 | 0.31 |
Mtus1 |
mitochondrial tumor suppressor 1 |
3762 |
0.18 |
| chr6_113062410_113062570 | 0.30 |
Gm22591 |
predicted gene, 22591 |
1792 |
0.2 |
| chr10_67056022_67056194 | 0.30 |
Reep3 |
receptor accessory protein 3 |
18786 |
0.17 |
| chr11_95252192_95252390 | 0.30 |
Tac4 |
tachykinin 4 |
9238 |
0.14 |
| chr11_98769689_98769972 | 0.30 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
596 |
0.57 |
| chr9_96116991_96117142 | 0.30 |
Gk5 |
glycerol kinase 5 (putative) |
2296 |
0.3 |
| chr2_110290286_110290470 | 0.30 |
Bbox1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
15363 |
0.21 |
| chr15_76695232_76695387 | 0.30 |
Gpt |
glutamic pyruvic transaminase, soluble |
407 |
0.62 |
| chr6_5517592_5517751 | 0.30 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
21362 |
0.26 |
| chr13_39657075_39657270 | 0.30 |
Gm47352 |
predicted gene, 47352 |
7661 |
0.21 |
| chr13_102646594_102646778 | 0.30 |
Gm47014 |
predicted gene, 47014 |
42153 |
0.14 |
| chr10_80405043_80405226 | 0.30 |
Uqcr11 |
ubiquinol-cytochrome c reductase, complex III subunit XI |
1632 |
0.16 |
| chr5_66328869_66329031 | 0.30 |
Gm43790 |
predicted gene 43790 |
5933 |
0.14 |
| chr11_94274186_94274507 | 0.30 |
Gm21885 |
predicted gene, 21885 |
7527 |
0.15 |
| chr15_59786780_59786984 | 0.29 |
Gm19510 |
predicted gene, 19510 |
8077 |
0.27 |
| chr2_173159311_173159586 | 0.29 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
6366 |
0.18 |
| chr1_165661837_165661988 | 0.29 |
Gm18407 |
predicted gene, 18407 |
17224 |
0.1 |
| chr16_24238329_24238511 | 0.29 |
Gm31814 |
predicted gene, 31814 |
21896 |
0.19 |
| chr18_56399298_56399460 | 0.29 |
Gramd3 |
GRAM domain containing 3 |
958 |
0.58 |
| chr11_97703209_97703365 | 0.29 |
Psmb3 |
proteasome (prosome, macropain) subunit, beta type 3 |
112 |
0.9 |
| chr1_121332590_121332782 | 0.29 |
Insig2 |
insulin induced gene 2 |
97 |
0.96 |
| chr10_84581307_84581543 | 0.29 |
Tcp11l2 |
t-complex 11 (mouse) like 2 |
4528 |
0.15 |
| chr6_51600845_51601052 | 0.29 |
Gm22914 |
predicted gene, 22914 |
20684 |
0.2 |
| chr2_84937644_84937820 | 0.29 |
Slc43a3 |
solute carrier family 43, member 3 |
842 |
0.5 |
| chr8_127184160_127184328 | 0.29 |
Pard3 |
par-3 family cell polarity regulator |
12840 |
0.27 |
| chr1_191628618_191628785 | 0.28 |
Gm37349 |
predicted gene, 37349 |
31054 |
0.13 |
| chr9_65237582_65237733 | 0.28 |
Parp16 |
poly (ADP-ribose) polymerase family, member 16 |
22503 |
0.1 |
| chr6_108660022_108660191 | 0.28 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
523 |
0.57 |
| chr5_118560730_118560903 | 0.28 |
Med13l |
mediator complex subunit 13-like |
77 |
0.96 |
| chr2_136986812_136986999 | 0.28 |
Slx4ip |
SLX4 interacting protein |
81362 |
0.08 |
| chr10_61383039_61383232 | 0.28 |
Pald1 |
phosphatase domain containing, paladin 1 |
395 |
0.75 |
| chr9_74528700_74528872 | 0.28 |
Gm28622 |
predicted gene 28622 |
32592 |
0.18 |
| chr11_102240756_102240907 | 0.28 |
Hrob |
homologous recombination factor with OB-fold |
8051 |
0.09 |
| chr4_8714834_8715372 | 0.28 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
4763 |
0.29 |
| chr2_24742080_24742251 | 0.28 |
Cacna1b |
calcium channel, voltage-dependent, N type, alpha 1B subunit |
20882 |
0.16 |
| chr1_184069803_184070002 | 0.28 |
Dusp10 |
dual specificity phosphatase 10 |
35521 |
0.18 |
| chr9_107657610_107657902 | 0.28 |
Slc38a3 |
solute carrier family 38, member 3 |
495 |
0.58 |
| chr9_20951552_20951703 | 0.28 |
Dnmt1 |
DNA methyltransferase (cytosine-5) 1 |
978 |
0.34 |
| chr3_133534210_133534372 | 0.28 |
Tet2 |
tet methylcytosine dioxygenase 2 |
9995 |
0.17 |
| chr16_95824094_95824245 | 0.28 |
2810404F17Rik |
RIKEN cDNA 2810404F17 gene |
15852 |
0.17 |
| chr8_119862106_119862265 | 0.28 |
Klhl36 |
kelch-like 36 |
81 |
0.97 |
| chr4_152010395_152010554 | 0.28 |
Gm9768 |
predicted gene 9768 |
855 |
0.38 |
| chr6_121992123_121992633 | 0.28 |
Mug2 |
murinoglobulin 2 |
14383 |
0.19 |
| chr11_120695849_120696008 | 0.27 |
Aspscr1 |
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
6586 |
0.06 |
| chr17_45691675_45691964 | 0.27 |
Mrpl14 |
mitochondrial ribosomal protein L14 |
3597 |
0.14 |
| chr12_17425859_17426047 | 0.27 |
Gm36752 |
predicted gene, 36752 |
10665 |
0.19 |
| chr2_25225323_25225479 | 0.27 |
Tubb4b |
tubulin, beta 4B class IVB |
699 |
0.33 |
| chr11_106122887_106123054 | 0.27 |
Gm11672 |
predicted gene 11672 |
4381 |
0.13 |
| chr1_171329168_171329338 | 0.27 |
Dedd |
death effector domain-containing |
108 |
0.89 |
| chr10_79936936_79937107 | 0.27 |
Arid3a |
AT rich interactive domain 3A (BRIGHT-like) |
6597 |
0.06 |
| chr2_167719609_167719764 | 0.27 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
28505 |
0.1 |
| chr1_72851799_72851950 | 0.27 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
3175 |
0.28 |
| chr12_82337746_82337908 | 0.27 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
4440 |
0.32 |
| chr6_87513163_87513315 | 0.27 |
Arhgap25 |
Rho GTPase activating protein 25 |
16925 |
0.14 |
| chr13_98594038_98594200 | 0.27 |
Gm4815 |
predicted gene 4815 |
19382 |
0.12 |
| chr19_10193896_10194210 | 0.27 |
Fads1 |
fatty acid desaturase 1 |
734 |
0.46 |
| chr10_77589222_77589404 | 0.27 |
Pttg1ip |
pituitary tumor-transforming 1 interacting protein |
81 |
0.94 |
| chr18_43250442_43250599 | 0.27 |
Stk32a |
serine/threonine kinase 32A |
7480 |
0.24 |
| chr2_144256907_144257089 | 0.27 |
Snx5 |
sorting nexin 5 |
1595 |
0.22 |
| chr2_137080581_137080763 | 0.26 |
Jag1 |
jagged 1 |
5277 |
0.31 |
| chr9_106453257_106453493 | 0.26 |
Pcbp4 |
poly(rC) binding protein 4 |
32 |
0.86 |
| chr11_95032963_95033336 | 0.26 |
Gm11513 |
predicted gene 11513 |
390 |
0.76 |
| chr6_121196988_121197205 | 0.26 |
Gm44014 |
predicted gene, 44014 |
1087 |
0.4 |
| chr8_4258328_4258479 | 0.26 |
Ctxn1 |
cortexin 1 |
871 |
0.34 |
| chr12_80218314_80218489 | 0.26 |
Gm47767 |
predicted gene, 47767 |
9158 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.4 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.5 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.3 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.2 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.2 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 0.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.0 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.6 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.2 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.2 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.2 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0014745 | negative regulation of muscle adaptation(GO:0014745) |
| 0.0 | 0.1 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.0 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.1 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.0 | GO:0051610 | serotonin uptake(GO:0051610) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.2 | 0.5 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.1 | 0.3 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0018641 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0052622 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0043910 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.6 | GO:0043765 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.9 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |