Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
Results for Xbp1_Creb3l1
Z-value: 1.06
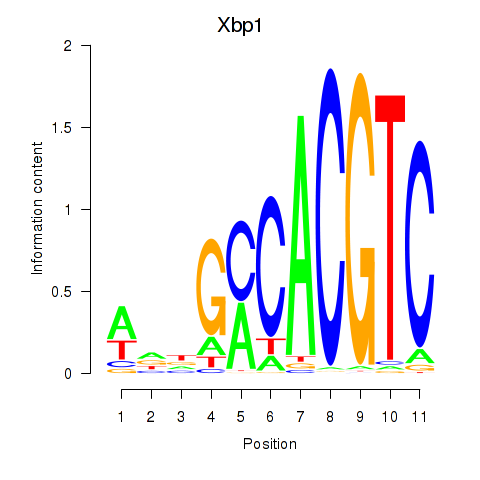
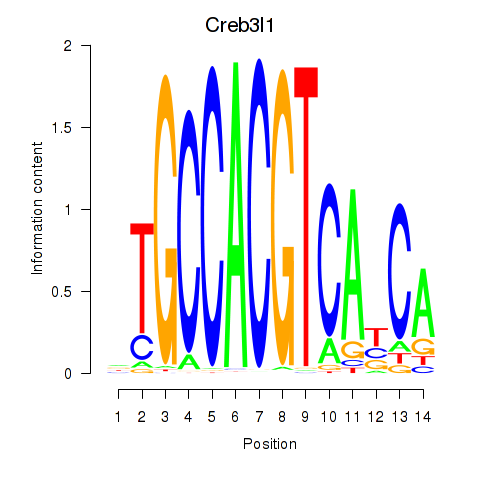
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Xbp1
|
ENSMUSG00000020484.12 | X-box binding protein 1 |
|
Creb3l1
|
ENSMUSG00000027230.9 | cAMP responsive element binding protein 3-like 1 |
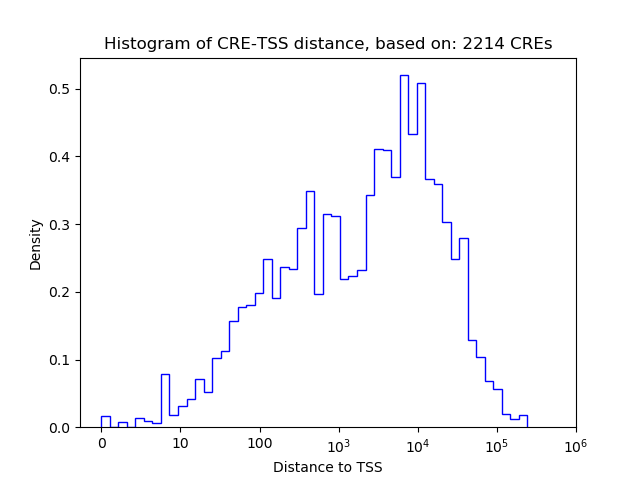
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_92008098_92008267 | Creb3l1 | 16320 | 0.133219 | 0.73 | 1.0e-01 | Click! |
| chr2_92004729_92004899 | Creb3l1 | 19688 | 0.126127 | 0.48 | 3.3e-01 | Click! |
| chr2_92008665_92009089 | Creb3l1 | 15625 | 0.134651 | -0.28 | 5.9e-01 | Click! |
| chr2_92012868_92013034 | Creb3l1 | 11551 | 0.143118 | -0.25 | 6.4e-01 | Click! |
| chr2_92027921_92028092 | Creb3l1 | 3504 | 0.191141 | -0.21 | 6.9e-01 | Click! |
| chr11_5529179_5529330 | Xbp1 | 7331 | 0.131215 | -0.72 | 1.1e-01 | Click! |
| chr11_5528014_5528184 | Xbp1 | 6176 | 0.135744 | -0.67 | 1.5e-01 | Click! |
| chr11_5523155_5523346 | Xbp1 | 1327 | 0.335220 | -0.62 | 1.9e-01 | Click! |
| chr11_5521552_5521734 | Xbp1 | 20 | 0.965863 | 0.58 | 2.3e-01 | Click! |
| chr11_5526021_5526577 | Xbp1 | 4376 | 0.149182 | -0.53 | 2.8e-01 | Click! |
Activity of the Xbp1_Creb3l1 motif across conditions
Conditions sorted by the z-value of the Xbp1_Creb3l1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
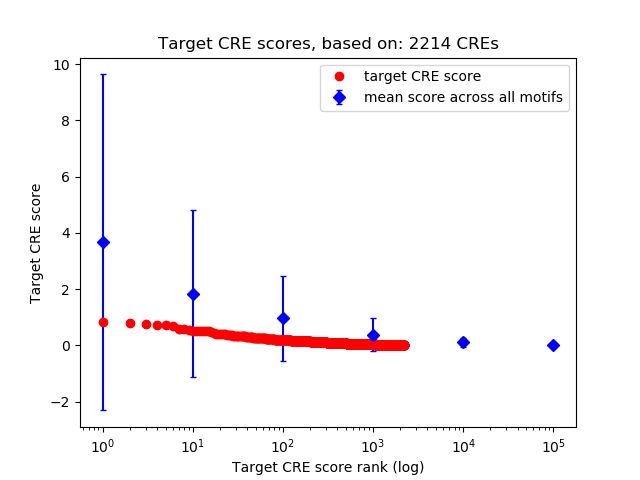
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_28899893_28900063 | 0.84 |
Gm31508 |
predicted gene, 31508 |
10251 |
0.18 |
| chr11_119191869_119192119 | 0.80 |
Gm11753 |
predicted gene 11753 |
7478 |
0.13 |
| chr6_118479189_118479349 | 0.76 |
Zfp9 |
zinc finger protein 9 |
51 |
0.97 |
| chr3_84214395_84214574 | 0.73 |
Trim2 |
tripartite motif-containing 2 |
1646 |
0.43 |
| chr11_86977739_86978000 | 0.72 |
Ypel2 |
yippee like 2 |
5845 |
0.19 |
| chr1_157506654_157506814 | 0.69 |
Sec16b |
SEC16 homolog B (S. cerevisiae) |
6 |
0.97 |
| chr17_50025214_50025398 | 0.59 |
AC133946.1 |
oxidoreductase NAD-binding domain containing 1 (OXNAD1) pseudogene |
52479 |
0.12 |
| chr15_86074327_86074478 | 0.58 |
Gramd4 |
GRAM domain containing 4 |
331 |
0.88 |
| chr9_124125701_124125852 | 0.56 |
Ccr5 |
chemokine (C-C motif) receptor 5 |
908 |
0.61 |
| chr6_128504617_128505108 | 0.52 |
Pzp |
PZP, alpha-2-macroglobulin like |
9639 |
0.09 |
| chr13_93625401_93625555 | 0.51 |
Gm15622 |
predicted gene 15622 |
96 |
0.96 |
| chr7_123196570_123196734 | 0.51 |
Tnrc6a |
trinucleotide repeat containing 6a |
16731 |
0.19 |
| chr8_105289865_105290239 | 0.51 |
Gm20163 |
predicted gene, 20163 |
63 |
0.64 |
| chr6_30563747_30563898 | 0.51 |
Cpa4 |
carboxypeptidase A4 |
4547 |
0.14 |
| chrX_164036094_164036254 | 0.51 |
Car5b |
carbonic anhydrase 5b, mitochondrial |
8177 |
0.19 |
| chr8_35432403_35432588 | 0.46 |
Gm34853 |
predicted gene, 34853 |
7239 |
0.18 |
| chr2_30407337_30407488 | 0.44 |
Crat |
carnitine acetyltransferase |
733 |
0.48 |
| chr12_8509811_8509989 | 0.42 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
1301 |
0.39 |
| chr8_123718352_123718527 | 0.41 |
6030466F02Rik |
RIKEN cDNA 6030466F02 gene |
15519 |
0.07 |
| chr7_4239400_4239603 | 0.40 |
Lilra5 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
1747 |
0.21 |
| chr4_123390896_123391047 | 0.40 |
Macf1 |
microtubule-actin crosslinking factor 1 |
433 |
0.8 |
| chr11_119089692_119089843 | 0.40 |
Cbx4 |
chromobox 4 |
3546 |
0.18 |
| chr2_172441299_172441621 | 0.40 |
Rtf2 |
replication termination factor 2 |
858 |
0.36 |
| chr4_131837128_131837286 | 0.38 |
Ptpru |
protein tyrosine phosphatase, receptor type, U |
1024 |
0.44 |
| chr5_8971094_8971245 | 0.37 |
Gm15610 |
predicted gene 15610 |
779 |
0.53 |
| chr1_39192284_39192607 | 0.35 |
Npas2 |
neuronal PAS domain protein 2 |
1286 |
0.45 |
| chr1_183369609_183369773 | 0.35 |
Mia3 |
melanoma inhibitory activity 3 |
138 |
0.92 |
| chr5_144885994_144886193 | 0.34 |
Smurf1 |
SMAD specific E3 ubiquitin protein ligase 1 |
383 |
0.83 |
| chr17_15192816_15193078 | 0.34 |
Gm35455 |
predicted gene, 35455 |
40788 |
0.13 |
| chr16_45377470_45377621 | 0.34 |
Cd200 |
CD200 antigen |
22767 |
0.16 |
| chr5_113735843_113736024 | 0.34 |
Ficd |
FIC domain containing |
130 |
0.93 |
| chr11_77793030_77793211 | 0.33 |
Gm10277 |
predicted gene 10277 |
5373 |
0.16 |
| chr9_61816682_61816855 | 0.33 |
Gm19208 |
predicted gene, 19208 |
34936 |
0.17 |
| chr9_57758758_57758995 | 0.33 |
Clk3 |
CDC-like kinase 3 |
3555 |
0.17 |
| chr15_101299611_101299792 | 0.32 |
Smim41 |
small integral membrane protein 41 |
6469 |
0.1 |
| chr18_75108255_75108425 | 0.32 |
Dym |
dymeclin |
6318 |
0.2 |
| chr8_41179352_41179538 | 0.32 |
Fgl1 |
fibrinogen-like protein 1 |
35708 |
0.13 |
| chr11_118454196_118454347 | 0.32 |
Gm11747 |
predicted gene 11747 |
724 |
0.56 |
| chr8_70200077_70200228 | 0.30 |
Slc25a42 |
solute carrier family 25, member 42 |
12104 |
0.09 |
| chr10_95408627_95408789 | 0.30 |
Socs2 |
suppressor of cytokine signaling 2 |
4026 |
0.16 |
| chr13_54575762_54575922 | 0.29 |
Arl10 |
ADP-ribosylation factor-like 10 |
795 |
0.43 |
| chr17_29496086_29496540 | 0.29 |
Pim1 |
proviral integration site 1 |
2906 |
0.15 |
| chr2_59600868_59601029 | 0.29 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
11094 |
0.23 |
| chr1_78568521_78568696 | 0.28 |
Mogat1 |
monoacylglycerol O-acyltransferase 1 |
30980 |
0.13 |
| chr9_7571293_7571444 | 0.28 |
Mmp27 |
matrix metallopeptidase 27 |
28 |
0.97 |
| chr11_115413035_115413484 | 0.28 |
Mrpl58 |
mitochondrial ribosomal protein L58 |
3099 |
0.1 |
| chr10_85103610_85103858 | 0.27 |
Tmem263 |
transmembrane protein 263 |
991 |
0.4 |
| chr2_6057072_6057245 | 0.27 |
Upf2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
6731 |
0.22 |
| chr12_86810853_86811037 | 0.27 |
Gm10095 |
predicted gene 10095 |
35522 |
0.14 |
| chr12_17376956_17377119 | 0.27 |
Mir3066 |
microRNA 3066 |
21645 |
0.13 |
| chr7_109562757_109562937 | 0.27 |
Denn2b |
DENN domain containing 2B |
22414 |
0.12 |
| chr3_96645795_96645997 | 0.26 |
Itga10 |
integrin, alpha 10 |
312 |
0.73 |
| chr14_51914351_51914597 | 0.26 |
Ndrg2 |
N-myc downstream regulated gene 2 |
316 |
0.76 |
| chr5_66014048_66014243 | 0.26 |
9130230L23Rik |
RIKEN cDNA 9130230L23 gene |
9860 |
0.12 |
| chr2_36082178_36082681 | 0.26 |
Lhx6 |
LIM homeobox protein 6 |
11844 |
0.12 |
| chr12_80434710_80434908 | 0.26 |
Gm30025 |
predicted gene, 30025 |
400 |
0.74 |
| chr4_108216816_108217079 | 0.26 |
Zyg11a |
zyg-11 family member A, cell cycle regulator |
975 |
0.5 |
| chr5_144891376_144891550 | 0.25 |
Smurf1 |
SMAD specific E3 ubiquitin protein ligase 1 |
4987 |
0.17 |
| chr12_21183267_21183628 | 0.25 |
AC156032.1 |
|
63876 |
0.08 |
| chr9_63755131_63755299 | 0.25 |
Smad3 |
SMAD family member 3 |
2779 |
0.31 |
| chr16_90856531_90856709 | 0.25 |
Gm36363 |
predicted gene, 36363 |
5657 |
0.14 |
| chr16_95729140_95729334 | 0.25 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
11503 |
0.18 |
| chr5_113918817_113918980 | 0.25 |
Gm22056 |
predicted gene, 22056 |
9906 |
0.11 |
| chr1_75661119_75661284 | 0.24 |
Gm5257 |
predicted gene 5257 |
24811 |
0.15 |
| chr3_93520535_93520702 | 0.24 |
S100a11 |
S100 calcium binding protein A11 |
130 |
0.93 |
| chr8_71366812_71367005 | 0.24 |
Use1 |
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
34 |
0.95 |
| chr5_114707898_114708063 | 0.23 |
Tchp |
trichoplein, keratin filament binding |
116 |
0.93 |
| chr2_156462736_156462911 | 0.22 |
Gm14224 |
predicted gene 14224 |
7172 |
0.12 |
| chr6_91117239_91117541 | 0.22 |
Nup210 |
nucleoporin 210 |
561 |
0.7 |
| chr15_85645356_85645520 | 0.22 |
Gm49539 |
predicted gene, 49539 |
645 |
0.66 |
| chr8_11727425_11727615 | 0.22 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
201 |
0.88 |
| chr6_145306818_145306981 | 0.22 |
Gm44428 |
predicted gene, 44428 |
2589 |
0.17 |
| chr4_144981227_144981590 | 0.22 |
Vps13d |
vacuolar protein sorting 13D |
2553 |
0.3 |
| chr16_52249852_52250207 | 0.21 |
Alcam |
activated leukocyte cell adhesion molecule |
19032 |
0.27 |
| chr19_43511661_43512096 | 0.21 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
3987 |
0.15 |
| chr10_53597848_53598018 | 0.21 |
Asf1a |
anti-silencing function 1A histone chaperone |
176 |
0.62 |
| chr10_84381402_84381782 | 0.21 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
10796 |
0.2 |
| chr3_52708989_52709165 | 0.21 |
Gm22798 |
predicted gene, 22798 |
13678 |
0.2 |
| chr1_36535791_36536054 | 0.21 |
Ankrd23 |
ankyrin repeat domain 23 |
183 |
0.87 |
| chr19_8966562_8966728 | 0.21 |
Eef1g |
eukaryotic translation elongation factor 1 gamma |
396 |
0.66 |
| chr3_27710416_27710593 | 0.21 |
Fndc3b |
fibronectin type III domain containing 3B |
65 |
0.98 |
| chr10_84409024_84409261 | 0.20 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
16754 |
0.17 |
| chr2_168742510_168742664 | 0.20 |
Atp9a |
ATPase, class II, type 9A |
178 |
0.95 |
| chr1_131975685_131975855 | 0.20 |
Slc45a3 |
solute carrier family 45, member 3 |
949 |
0.44 |
| chr4_61439599_61439764 | 0.20 |
Mup15 |
major urinary protein 15 |
62 |
0.97 |
| chr10_84803969_84804150 | 0.20 |
Gm24226 |
predicted gene, 24226 |
7124 |
0.23 |
| chr3_153921648_153921917 | 0.20 |
Acadm |
acyl-Coenzyme A dehydrogenase, medium chain |
8360 |
0.09 |
| chr4_135434067_135434419 | 0.20 |
Rcan3 |
regulator of calcineurin 3 |
390 |
0.72 |
| chr2_104035717_104035881 | 0.20 |
Gm24644 |
predicted gene, 24644 |
4870 |
0.12 |
| chr15_99847019_99847476 | 0.20 |
Lima1 |
LIM domain and actin binding 1 |
27104 |
0.07 |
| chr9_47637935_47638086 | 0.20 |
Cadm1 |
cell adhesion molecule 1 |
107637 |
0.06 |
| chr11_86807573_86807779 | 0.20 |
Dhx40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
70 |
0.98 |
| chr13_64153328_64153492 | 0.20 |
Zfp367 |
zinc finger protein 367 |
208 |
0.83 |
| chr4_83221216_83221400 | 0.19 |
Ttc39b |
tetratricopeptide repeat domain 39B |
11862 |
0.19 |
| chr4_61595719_61595883 | 0.19 |
Mup17 |
major urinary protein 17 |
70 |
0.97 |
| chr19_3695153_3695350 | 0.19 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
8687 |
0.1 |
| chr17_12371689_12371855 | 0.19 |
Plg |
plasminogen |
6836 |
0.16 |
| chr7_44869987_44870160 | 0.19 |
Ptov1 |
prostate tumor over expressed gene 1 |
285 |
0.74 |
| chr4_60139744_60139909 | 0.19 |
Mup2 |
major urinary protein 2 |
31 |
0.97 |
| chr3_139205794_139206161 | 0.19 |
Stpg2 |
sperm tail PG rich repeat containing 2 |
84 |
0.98 |
| chr11_17745312_17745477 | 0.19 |
Gm12016 |
predicted gene 12016 |
106211 |
0.07 |
| chr13_94143352_94143530 | 0.19 |
Gm15907 |
predicted gene 15907 |
3324 |
0.24 |
| chr1_171294216_171294434 | 0.18 |
Ufc1 |
ubiquitin-fold modifier conjugating enzyme 1 |
615 |
0.44 |
| chr11_59428409_59428580 | 0.18 |
Snap47 |
synaptosomal-associated protein, 47 |
18997 |
0.1 |
| chr6_55001492_55001666 | 0.18 |
Ggct |
gamma-glutamyl cyclotransferase |
8629 |
0.17 |
| chr5_117245335_117245486 | 0.18 |
Taok3 |
TAO kinase 3 |
4950 |
0.16 |
| chr4_61519388_61519546 | 0.18 |
Mup16 |
major urinary protein 16 |
0 |
0.98 |
| chr4_153957077_153957238 | 0.18 |
A430005L14Rik |
RIKEN cDNA A430005L14 gene |
80 |
0.95 |
| chr15_85036576_85036766 | 0.18 |
Fam118a |
family with sequence similarity 118, member A |
418 |
0.76 |
| chr12_28910610_28910780 | 0.18 |
Gm31508 |
predicted gene, 31508 |
466 |
0.82 |
| chr5_30231710_30232189 | 0.18 |
Selenoi |
selenoprotein I |
632 |
0.63 |
| chr4_44991523_44991952 | 0.18 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
10237 |
0.11 |
| chr7_45139988_45140693 | 0.18 |
Flt3l |
FMS-like tyrosine kinase 3 ligand |
3908 |
0.05 |
| chr17_74716795_74716985 | 0.18 |
Ttc27 |
tetratricopeptide repeat domain 27 |
842 |
0.65 |
| chr12_105297953_105298115 | 0.18 |
Tunar |
Tcl1 upstream neural differentiation associated RNA |
35836 |
0.14 |
| chr8_127295141_127295294 | 0.18 |
Pard3 |
par-3 family cell polarity regulator |
4368 |
0.35 |
| chr18_34331012_34331178 | 0.18 |
Srp19 |
signal recognition particle 19 |
28 |
0.98 |
| chr8_33987157_33987321 | 0.18 |
Gm45817 |
predicted gene 45817 |
39 |
0.96 |
| chr18_61489282_61489444 | 0.17 |
Gm46637 |
predicted gene, 46637 |
4597 |
0.14 |
| chr5_74943885_74944036 | 0.17 |
Gm6116 |
predicted gene 6116 |
5230 |
0.2 |
| chr4_60662258_60662429 | 0.17 |
Mup11 |
major urinary protein 11 |
15 |
0.97 |
| chr19_5770906_5771201 | 0.17 |
Scyl1 |
SCY1-like 1 (S. cerevisiae) |
106 |
0.91 |
| chr11_58093075_58093271 | 0.17 |
Gm12248 |
predicted gene 12248 |
39 |
0.96 |
| chr1_133278743_133278947 | 0.17 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
4591 |
0.15 |
| chr16_97418050_97418216 | 0.17 |
Bace2 |
beta-site APP-cleaving enzyme 2 |
3055 |
0.21 |
| chr5_66074323_66074506 | 0.17 |
Gm43775 |
predicted gene 43775 |
1519 |
0.29 |
| chr17_28930275_28930478 | 0.17 |
Gm16191 |
predicted gene 16191 |
4045 |
0.1 |
| chr4_137468313_137468498 | 0.17 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
364 |
0.81 |
| chr2_127506298_127506464 | 0.17 |
AL731831.1 |
prominin 2 (Prom2), pseudogene |
12914 |
0.13 |
| chr10_128980928_128981107 | 0.17 |
Olfr9 |
olfactory receptor 9 |
8414 |
0.09 |
| chr11_21428880_21429060 | 0.17 |
Gm12044 |
predicted gene 12044 |
22384 |
0.14 |
| chr1_38864480_38864651 | 0.16 |
Chst10 |
carbohydrate sulfotransferase 10 |
7195 |
0.16 |
| chr19_46133855_46134398 | 0.16 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
2229 |
0.19 |
| chr12_4039052_4039225 | 0.16 |
Efr3b |
EFR3 homolog B |
223 |
0.92 |
| chr9_73039846_73040053 | 0.16 |
Pigbos1 |
Pigb opposite strand 1 |
219 |
0.5 |
| chr7_27331182_27331346 | 0.16 |
Ltbp4 |
latent transforming growth factor beta binding protein 4 |
2349 |
0.17 |
| chr14_77343821_77343990 | 0.16 |
Gm49001 |
predicted gene, 49001 |
281 |
0.93 |
| chr4_60070204_60070385 | 0.16 |
Mup7 |
major urinary protein 7 |
117 |
0.96 |
| chr12_79684888_79685267 | 0.16 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
239656 |
0.02 |
| chr7_44999841_45000005 | 0.16 |
Irf3 |
interferon regulatory factor 3 |
244 |
0.73 |
| chr2_168127857_168128015 | 0.16 |
AL831766.1 |
breast carcinoma amplified sequence 4 (BCAS4) pseudogene |
13059 |
0.12 |
| chr13_12454823_12454987 | 0.16 |
Lgals8 |
lectin, galactose binding, soluble 8 |
109 |
0.96 |
| chr2_148040189_148040578 | 0.16 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
180 |
0.94 |
| chr14_17683673_17683824 | 0.16 |
Thrb |
thyroid hormone receptor beta |
22852 |
0.26 |
| chr2_156732212_156732375 | 0.16 |
Dlgap4 |
DLG associated protein 4 |
10801 |
0.13 |
| chr2_45253993_45254149 | 0.16 |
Gm13475 |
predicted gene 13475 |
75161 |
0.1 |
| chr3_27686853_27687185 | 0.16 |
Fndc3b |
fibronectin type III domain containing 3B |
23379 |
0.24 |
| chr5_9047464_9048021 | 0.16 |
Gm40264 |
predicted gene, 40264 |
12618 |
0.15 |
| chr4_60222408_60222586 | 0.16 |
Mup8 |
major urinary protein 8 |
83 |
0.97 |
| chr6_12466411_12466591 | 0.16 |
Thsd7a |
thrombospondin, type I, domain containing 7A |
47140 |
0.18 |
| chr16_4753759_4754349 | 0.16 |
Hmox2 |
heme oxygenase 2 |
2791 |
0.17 |
| chr6_5190649_5191077 | 0.16 |
Pon1 |
paraoxonase 1 |
2900 |
0.26 |
| chr15_88894079_88894230 | 0.16 |
Gm20621 |
predicted gene 20621 |
2629 |
0.2 |
| chr8_70594556_70594707 | 0.16 |
Isyna1 |
myo-inositol 1-phosphate synthase A1 |
96 |
0.93 |
| chr1_152556824_152557014 | 0.15 |
Rgl1 |
ral guanine nucleotide dissociation stimulator,-like 1 |
3875 |
0.31 |
| chr10_99615923_99616079 | 0.15 |
Gm20110 |
predicted gene, 20110 |
6830 |
0.19 |
| chr16_50430297_50430475 | 0.15 |
Bbx |
bobby sox HMG box containing |
474 |
0.88 |
| chr7_16924149_16924320 | 0.15 |
Calm3 |
calmodulin 3 |
120 |
0.92 |
| chr5_138979896_138980396 | 0.15 |
Pdgfa |
platelet derived growth factor, alpha |
14135 |
0.17 |
| chr4_60501802_60501955 | 0.15 |
Mup1 |
major urinary protein 1 |
25 |
0.94 |
| chr4_32520693_32520870 | 0.15 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
19276 |
0.19 |
| chr4_116685113_116685395 | 0.15 |
Prdx1 |
peroxiredoxin 1 |
290 |
0.83 |
| chr5_129501132_129501293 | 0.15 |
Sfswap |
splicing factor SWAP |
9 |
0.74 |
| chr6_121329980_121330165 | 0.15 |
Gm24855 |
predicted gene, 24855 |
4396 |
0.17 |
| chr5_45476407_45476609 | 0.15 |
Lap3 |
leucine aminopeptidase 3 |
16866 |
0.11 |
| chr8_3631331_3631522 | 0.15 |
Stxbp2 |
syntaxin binding protein 2 |
256 |
0.79 |
| chr5_116453577_116453751 | 0.15 |
Srrm4 |
serine/arginine repetitive matrix 4 |
249 |
0.88 |
| chr2_109978777_109979106 | 0.15 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
3399 |
0.26 |
| chr8_10899998_10900419 | 0.15 |
4833411C07Rik |
RIKEN cDNA 4833411C07 gene |
271 |
0.54 |
| chr6_28480072_28480271 | 0.15 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
177 |
0.93 |
| chr8_27174337_27174510 | 0.15 |
Rab11fip1 |
RAB11 family interacting protein 1 (class I) |
204 |
0.88 |
| chr9_69289046_69289527 | 0.15 |
Rora |
RAR-related orphan receptor alpha |
396 |
0.89 |
| chr9_111211971_111212321 | 0.15 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
2047 |
0.3 |
| chr8_77098217_77098409 | 0.15 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
29700 |
0.19 |
| chr10_59504215_59504400 | 0.15 |
Mcu |
mitochondrial calcium uniporter |
46147 |
0.13 |
| chr9_61596903_61597054 | 0.15 |
Gm34424 |
predicted gene, 34424 |
63563 |
0.12 |
| chr10_13967469_13967637 | 0.15 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
529 |
0.78 |
| chr16_92415881_92416059 | 0.15 |
Rcan1 |
regulator of calcineurin 1 |
15893 |
0.13 |
| chr3_31122977_31123199 | 0.14 |
Skil |
SKI-like |
26255 |
0.16 |
| chr11_83834151_83834516 | 0.14 |
Gm12576 |
predicted gene 12576 |
15160 |
0.12 |
| chr4_118137948_118138112 | 0.14 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
3116 |
0.21 |
| chr1_133452688_133453146 | 0.14 |
Sox13 |
SRY (sex determining region Y)-box 13 |
28540 |
0.16 |
| chr7_30360114_30360296 | 0.14 |
Lrfn3 |
leucine rich repeat and fibronectin type III domain containing 3 |
2567 |
0.1 |
| chr19_43512283_43512443 | 0.14 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
3502 |
0.15 |
| chr10_18056009_18056160 | 0.14 |
Reps1 |
RalBP1 associated Eps domain containing protein |
80 |
0.97 |
| chr14_122104924_122105095 | 0.14 |
A330035P11Rik |
RIKEN cDNA A330035P11 gene |
1944 |
0.21 |
| chr16_21646037_21646193 | 0.14 |
Vps8 |
VPS8 CORVET complex subunit |
18484 |
0.2 |
| chr4_53023825_53023983 | 0.14 |
Nipsnap3b |
nipsnap homolog 3B |
2928 |
0.22 |
| chr11_19992826_19993027 | 0.14 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
38356 |
0.18 |
| chr7_73574787_73575110 | 0.14 |
1810026B05Rik |
RIKEN cDNA 1810026B05 gene |
16553 |
0.1 |
| chr4_118158547_118158710 | 0.14 |
Kdm4a |
lysine (K)-specific demethylase 4A |
3029 |
0.2 |
| chr7_100042239_100042447 | 0.14 |
Chrdl2 |
chordin-like 2 |
20244 |
0.14 |
| chr2_150786904_150787055 | 0.14 |
Pygb |
brain glycogen phosphorylase |
244 |
0.91 |
| chr5_112893601_112893976 | 0.14 |
Myo18b |
myosin XVIIIb |
2574 |
0.29 |
| chr6_121841280_121841702 | 0.14 |
Mug1 |
murinoglobulin 1 |
387 |
0.88 |
| chr18_12213171_12213322 | 0.14 |
Npc1 |
NPC intracellular cholesterol transporter 1 |
692 |
0.62 |
| chr9_103471973_103472146 | 0.14 |
Gm16252 |
predicted gene 16252 |
25 |
0.96 |
| chr3_83032424_83032575 | 0.14 |
Fga |
fibrinogen alpha chain |
6284 |
0.15 |
| chr1_183367200_183367450 | 0.14 |
Mia3 |
melanoma inhibitory activity 3 |
2228 |
0.21 |
| chr5_139499620_139499800 | 0.14 |
Zfand2a |
zinc finger, AN1-type domain 2A |
15161 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.3 | GO:0052042 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.4 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.0 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0061623 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.0 | GO:2000591 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.2 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |