Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
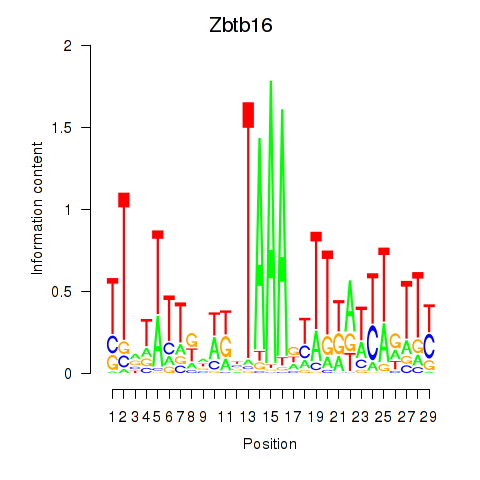
Results for Zbtb16
Z-value: 1.57
Transcription factors associated with Zbtb16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb16
|
ENSMUSG00000066687.4 | zinc finger and BTB domain containing 16 |
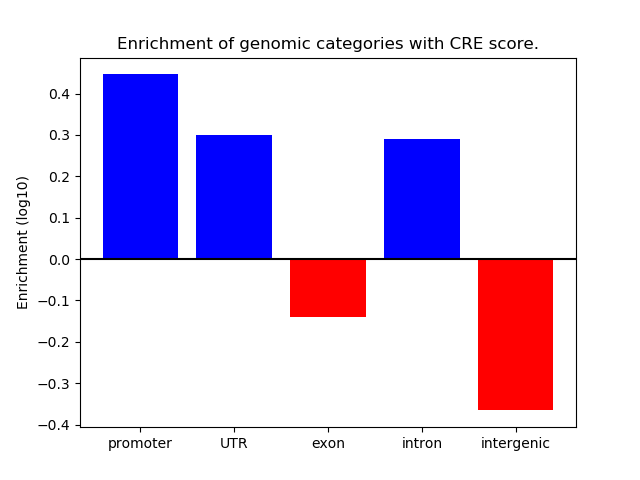
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_48835896_48836064 | Zbtb16 | 35 | 0.982436 | 0.95 | 4.2e-03 | Click! |
| chr9_48784989_48785185 | Zbtb16 | 50858 | 0.146076 | 0.82 | 4.6e-02 | Click! |
| chr9_48835645_48835854 | Zbtb16 | 196 | 0.956505 | 0.81 | 4.9e-02 | Click! |
| chr9_48836417_48836585 | Zbtb16 | 279 | 0.930627 | 0.77 | 7.4e-02 | Click! |
| chr9_48740241_48740405 | Zbtb16 | 95622 | 0.071587 | 0.74 | 9.1e-02 | Click! |
Activity of the Zbtb16 motif across conditions
Conditions sorted by the z-value of the Zbtb16 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
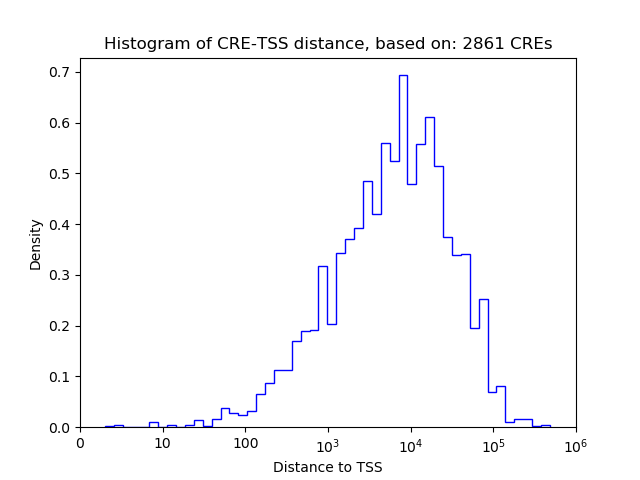
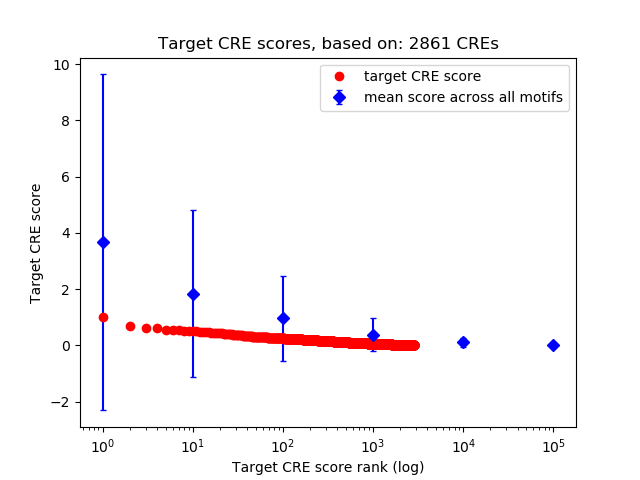
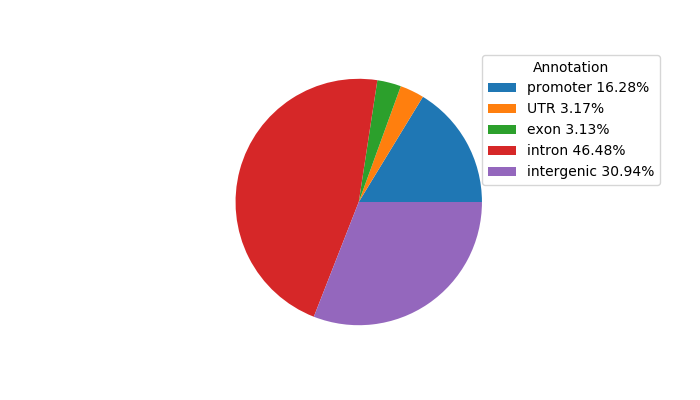
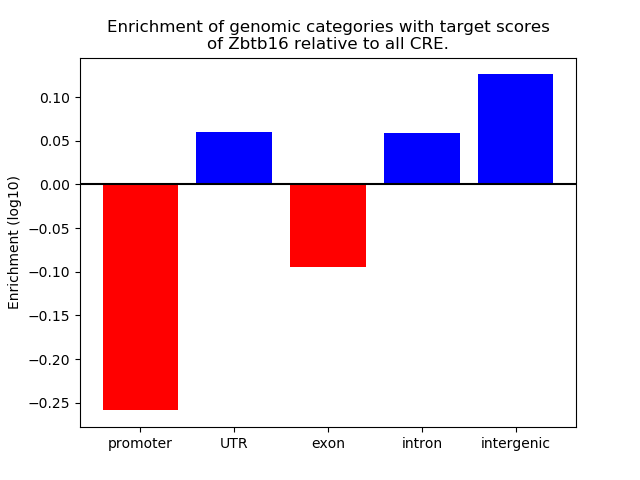
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_61942367_61942518 | 1.01 |
Gm37947 |
predicted gene, 37947 |
40860 |
0.16 |
| chr8_93397874_93398136 | 0.67 |
Gm45764 |
predicted gene 45764 |
900 |
0.48 |
| chr5_9079887_9080138 | 0.62 |
Tmem243 |
transmembrane protein 243, mitochondrial |
20656 |
0.14 |
| chr6_33039327_33039482 | 0.60 |
Gm14540 |
predicted gene 14540 |
936 |
0.53 |
| chr6_42222010_42222363 | 0.53 |
Tas2r144 |
taste receptor, type 2, member 144 |
6858 |
0.13 |
| chr11_102240756_102240907 | 0.53 |
Hrob |
homologous recombination factor with OB-fold |
8051 |
0.09 |
| chr12_79967016_79967483 | 0.53 |
Gm8275 |
predicted gene 8275 |
12602 |
0.18 |
| chr15_10997104_10997270 | 0.52 |
Slc45a2 |
solute carrier family 45, member 2 |
3534 |
0.2 |
| chr19_23110526_23110678 | 0.51 |
2410080I02Rik |
RIKEN cDNA 2410080I02 gene |
24298 |
0.14 |
| chr8_10939586_10939983 | 0.50 |
Gm44955 |
predicted gene 44955 |
8106 |
0.11 |
| chr11_121616042_121616235 | 0.50 |
Tbcd |
tubulin-specific chaperone d |
14433 |
0.2 |
| chr11_8607022_8607173 | 0.47 |
Tns3 |
tensin 3 |
17790 |
0.28 |
| chr1_106813159_106813457 | 0.46 |
Vps4b |
vacuolar protein sorting 4B |
16580 |
0.18 |
| chr6_42254630_42254929 | 0.46 |
Tmem139 |
transmembrane protein 139 |
7180 |
0.11 |
| chr3_83002544_83002701 | 0.46 |
Fgg |
fibrinogen gamma chain |
5102 |
0.16 |
| chr1_165609107_165609258 | 0.45 |
Mpzl1 |
myelin protein zero-like 1 |
364 |
0.78 |
| chr2_90518427_90518625 | 0.45 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
39276 |
0.14 |
| chr6_108648533_108648699 | 0.44 |
Gm17055 |
predicted gene 17055 |
8301 |
0.16 |
| chr12_109991265_109991416 | 0.44 |
Gm34667 |
predicted gene, 34667 |
32533 |
0.1 |
| chr1_138606335_138606734 | 0.43 |
Nek7 |
NIMA (never in mitosis gene a)-related expressed kinase 7 |
13140 |
0.17 |
| chr16_30628279_30628596 | 0.42 |
Fam43a |
family with sequence similarity 43, member A |
28714 |
0.17 |
| chr17_32493234_32493405 | 0.42 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
1161 |
0.38 |
| chr13_34793448_34793846 | 0.42 |
Gm47157 |
predicted gene, 47157 |
20897 |
0.13 |
| chr11_62487235_62487386 | 0.42 |
Gm12278 |
predicted gene 12278 |
4513 |
0.13 |
| chr16_71739508_71739663 | 0.40 |
Gm22797 |
predicted gene, 22797 |
75797 |
0.11 |
| chr6_128146811_128146971 | 0.39 |
Tspan9 |
tetraspanin 9 |
3297 |
0.15 |
| chr7_7187642_7187793 | 0.39 |
Zfp418 |
zinc finger protein 418 |
16363 |
0.1 |
| chr19_46899287_46899584 | 0.38 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
10140 |
0.16 |
| chr3_41582728_41583000 | 0.37 |
Jade1 |
jade family PHD finger 1 |
2008 |
0.28 |
| chr13_64126684_64127007 | 0.37 |
Slc35d2 |
solute carrier family 35, member D2 |
2460 |
0.2 |
| chr4_100479215_100479588 | 0.37 |
Ube2u |
ubiquitin-conjugating enzyme E2U (putative) |
534 |
0.82 |
| chr5_136379407_136379558 | 0.36 |
Mir721 |
microRNA 721 |
3679 |
0.2 |
| chr2_12867074_12867267 | 0.35 |
Pter |
phosphotriesterase related |
56871 |
0.13 |
| chr2_152663214_152663365 | 0.35 |
H13 |
histocompatibility 13 |
6172 |
0.1 |
| chr4_6885340_6885491 | 0.35 |
Tox |
thymocyte selection-associated high mobility group box |
105068 |
0.07 |
| chr14_114348500_114348688 | 0.35 |
Gm19829 |
predicted gene, 19829 |
37791 |
0.23 |
| chr10_24131559_24132178 | 0.35 |
Gm40614 |
predicted gene, 40614 |
612 |
0.59 |
| chr19_30128744_30128991 | 0.34 |
Gldc |
glycine decarboxylase |
16364 |
0.18 |
| chr6_147064387_147064568 | 0.34 |
Mrps35 |
mitochondrial ribosomal protein S35 |
3226 |
0.17 |
| chr10_8089089_8089240 | 0.34 |
Gm48614 |
predicted gene, 48614 |
67872 |
0.11 |
| chr11_35179112_35179275 | 0.33 |
Slit3 |
slit guidance ligand 3 |
1410 |
0.51 |
| chr15_38070580_38070794 | 0.33 |
Ubr5 |
ubiquitin protein ligase E3 component n-recognin 5 |
7738 |
0.16 |
| chr9_4380683_4380845 | 0.33 |
Msantd4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
2771 |
0.26 |
| chr2_23082221_23082376 | 0.32 |
Acbd5 |
acyl-Coenzyme A binding domain containing 5 |
1653 |
0.36 |
| chr16_76398590_76398917 | 0.31 |
Gm9843 |
predicted gene 9843 |
4899 |
0.21 |
| chr2_128713314_128713603 | 0.31 |
Mertk |
MER proto-oncogene tyrosine kinase |
14502 |
0.15 |
| chr11_70522908_70523209 | 0.31 |
Psmb6 |
proteasome (prosome, macropain) subunit, beta type 6 |
2309 |
0.1 |
| chr19_41812412_41812564 | 0.30 |
Arhgap19 |
Rho GTPase activating protein 19 |
10441 |
0.15 |
| chr5_137679989_137680170 | 0.30 |
Agfg2 |
ArfGAP with FG repeats 2 |
4601 |
0.09 |
| chr13_74897511_74897662 | 0.30 |
Hnrnpa1l2-ps |
heterogeneous nuclear ribonucleoprotein A1-like 2, pseudogene |
33281 |
0.19 |
| chr7_97622705_97622904 | 0.30 |
Rsf1os1 |
remodeling and spacing factor 1, opposite strand 1 |
22308 |
0.1 |
| chr10_68444703_68445037 | 0.30 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
80897 |
0.09 |
| chr12_8643213_8643385 | 0.29 |
Pum2 |
pumilio RNA-binding family member 2 |
30835 |
0.16 |
| chr5_87335535_87335718 | 0.29 |
Ugt2a3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
1569 |
0.26 |
| chr17_29496086_29496540 | 0.29 |
Pim1 |
proviral integration site 1 |
2906 |
0.15 |
| chr5_148965317_148966102 | 0.29 |
1810059H22Rik |
RIKEN cDNA 1810059H22 gene |
1435 |
0.22 |
| chr11_103897919_103898107 | 0.29 |
Gm8738 |
predicted gene 8738 |
17770 |
0.18 |
| chr16_23497586_23497756 | 0.29 |
Gm49514 |
predicted gene, 49514 |
16599 |
0.13 |
| chr11_101987341_101987516 | 0.28 |
Cfap97d1 |
CFAP97 domain containing 1 |
372 |
0.46 |
| chr16_30953629_30953810 | 0.28 |
Gm46565 |
predicted gene, 46565 |
18090 |
0.15 |
| chr16_30941233_30941532 | 0.28 |
Gm46565 |
predicted gene, 46565 |
5753 |
0.18 |
| chr12_52015542_52015693 | 0.28 |
Dtd2 |
D-tyrosyl-tRNA deacylase 2 |
9116 |
0.16 |
| chr11_111605518_111605669 | 0.28 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr10_52870918_52871110 | 0.28 |
Gm25664 |
predicted gene, 25664 |
34624 |
0.16 |
| chr10_71282973_71283124 | 0.28 |
Ube2d1 |
ubiquitin-conjugating enzyme E2D 1 |
2170 |
0.21 |
| chr8_83971032_83971407 | 0.28 |
Prkaca |
protein kinase, cAMP dependent, catalytic, alpha |
1774 |
0.16 |
| chr16_24394028_24394581 | 0.28 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr5_146270914_146271065 | 0.28 |
Cdk8 |
cyclin-dependent kinase 8 |
13196 |
0.14 |
| chr13_34313351_34313741 | 0.28 |
Gm47086 |
predicted gene, 47086 |
5262 |
0.21 |
| chr10_10580939_10581232 | 0.27 |
Rab32 |
RAB32, member RAS oncogene family |
22820 |
0.17 |
| chr5_123376677_123377005 | 0.27 |
5830487J09Rik |
RIKEN cDNA 5830487J09 gene |
9842 |
0.08 |
| chr1_170640374_170640534 | 0.27 |
Olfml2b |
olfactomedin-like 2B |
4078 |
0.21 |
| chr13_93889949_93890100 | 0.27 |
Gm25534 |
predicted gene, 25534 |
33287 |
0.15 |
| chr5_17814871_17815031 | 0.27 |
Cd36 |
CD36 molecule |
20745 |
0.27 |
| chr7_49761513_49761692 | 0.27 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
2449 |
0.31 |
| chr11_85738593_85738744 | 0.27 |
Bcas3os1 |
breast carcinoma amplified sequence 3, opposite strand 1 |
18924 |
0.14 |
| chr13_100525246_100525436 | 0.27 |
Ocln |
occludin |
27119 |
0.1 |
| chr13_60173158_60174110 | 0.26 |
Gm48488 |
predicted gene, 48488 |
3048 |
0.21 |
| chr2_33444795_33444946 | 0.26 |
Gm13536 |
predicted gene 13536 |
1932 |
0.27 |
| chr10_44699857_44700038 | 0.26 |
Gm47388 |
predicted gene, 47388 |
2767 |
0.17 |
| chr9_44127033_44127184 | 0.26 |
Mcam |
melanoma cell adhesion molecule |
7361 |
0.06 |
| chr10_68352578_68352735 | 0.26 |
4930545H06Rik |
RIKEN cDNA 4930545H06 gene |
31514 |
0.18 |
| chr13_12550644_12550996 | 0.26 |
Gm30239 |
predicted gene, 30239 |
7420 |
0.14 |
| chr6_50176151_50176315 | 0.26 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
4067 |
0.27 |
| chr19_10051741_10051938 | 0.26 |
Fads3 |
fatty acid desaturase 3 |
1112 |
0.39 |
| chr15_29829703_29829875 | 0.26 |
Gm23033 |
predicted gene, 23033 |
58685 |
0.14 |
| chr8_119431731_119431882 | 0.26 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
2318 |
0.24 |
| chr6_147064062_147064225 | 0.25 |
Mrps35 |
mitochondrial ribosomal protein S35 |
2892 |
0.18 |
| chr2_69228328_69228964 | 0.25 |
G6pc2 |
glucose-6-phosphatase, catalytic, 2 |
8618 |
0.15 |
| chr14_20319722_20320198 | 0.25 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
8531 |
0.12 |
| chr8_45255840_45256130 | 0.25 |
F11 |
coagulation factor XI |
6046 |
0.19 |
| chr5_72365932_72366096 | 0.25 |
Corin |
corin, serine peptidase |
5315 |
0.18 |
| chr5_129726157_129726319 | 0.25 |
Gm15903 |
predicted gene 15903 |
808 |
0.39 |
| chr11_50189181_50189338 | 0.25 |
Sqstm1 |
sequestosome 1 |
13695 |
0.1 |
| chr16_76225574_76225868 | 0.25 |
Gm26915 |
predicted gene, 26915 |
80736 |
0.08 |
| chr6_137466080_137466336 | 0.25 |
Ptpro |
protein tyrosine phosphatase, receptor type, O |
12180 |
0.22 |
| chr6_82133100_82133272 | 0.25 |
Gm15864 |
predicted gene 15864 |
80605 |
0.08 |
| chr16_31996383_31996610 | 0.25 |
Hmgb1-ps6 |
high mobility group box 1, pseudogene 6 |
4628 |
0.09 |
| chr18_46748548_46748699 | 0.24 |
Ap3s1 |
adaptor-related protein complex 3, sigma 1 subunit |
6681 |
0.12 |
| chr12_40508704_40509084 | 0.24 |
Dock4 |
dedicator of cytokinesis 4 |
62558 |
0.12 |
| chr14_47205203_47205370 | 0.24 |
Gm49136 |
predicted gene, 49136 |
5416 |
0.11 |
| chr8_35349975_35350150 | 0.24 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
25677 |
0.15 |
| chr1_93029762_93029954 | 0.24 |
Kif1a |
kinesin family member 1A |
2653 |
0.19 |
| chr12_83981935_83982155 | 0.24 |
Heatr4 |
HEAT repeat containing 4 |
2807 |
0.14 |
| chr4_143412215_143412366 | 0.24 |
Pramef8 |
PRAME family member 8 |
136 |
0.93 |
| chr2_38142003_38142233 | 0.24 |
Dennd1a |
DENN/MADD domain containing 1A |
23963 |
0.17 |
| chr2_172473776_172473949 | 0.24 |
Fam209 |
family with sequence similarity 209 |
1342 |
0.34 |
| chr10_71343007_71343170 | 0.24 |
Cisd1 |
CDGSH iron sulfur domain 1 |
1866 |
0.25 |
| chr15_11190225_11190388 | 0.24 |
Adamts12 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 |
125488 |
0.05 |
| chr2_156145846_156146301 | 0.24 |
Romo1 |
reactive oxygen species modulator 1 |
1833 |
0.18 |
| chr18_20210155_20210348 | 0.23 |
Dsg1c |
desmoglein 1 gamma |
37089 |
0.18 |
| chr7_105818652_105818813 | 0.23 |
Gm45912 |
predicted gene 45912 |
5074 |
0.12 |
| chr15_25677510_25677678 | 0.23 |
Myo10 |
myosin X |
3084 |
0.24 |
| chr11_87449212_87449363 | 0.23 |
Rnu3b3 |
U3B small nuclear RNA 3 |
660 |
0.48 |
| chr12_24614688_24615076 | 0.23 |
Gm6969 |
predicted pseudogene 6969 |
24128 |
0.13 |
| chr10_51722416_51722567 | 0.23 |
Rfx6 |
regulatory factor X, 6 |
28720 |
0.12 |
| chr4_146234816_146235138 | 0.23 |
Gm13169 |
predicted gene 13169 |
4977 |
0.13 |
| chr4_3875399_3875796 | 0.23 |
Mos |
Moloney sarcoma oncogene |
3492 |
0.17 |
| chr3_81995220_81995384 | 0.23 |
Asic5 |
acid-sensing (proton-gated) ion channel family member 5 |
1620 |
0.35 |
| chr14_59444898_59445049 | 0.23 |
Cab39l |
calcium binding protein 39-like |
3992 |
0.18 |
| chr9_59019307_59019466 | 0.23 |
Neo1 |
neogenin |
17039 |
0.22 |
| chr9_66330345_66330505 | 0.22 |
Herc1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
20025 |
0.18 |
| chr5_124376554_124376745 | 0.22 |
Sbno1 |
strawberry notch 1 |
8160 |
0.11 |
| chr12_85809600_85809751 | 0.22 |
Erg28 |
ergosterol biosynthesis 28 |
12575 |
0.16 |
| chr17_77596575_77596938 | 0.22 |
Gm17538 |
predicted gene, 17538 |
77944 |
0.11 |
| chr8_93146655_93146831 | 0.22 |
Gm45910 |
predicted gene 45910 |
7373 |
0.14 |
| chr6_108233581_108233762 | 0.22 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
19287 |
0.22 |
| chr1_49570933_49571107 | 0.22 |
Gm28388 |
predicted gene 28388 |
8566 |
0.29 |
| chr3_82316899_82317195 | 0.22 |
Map9 |
microtubule-associated protein 9 |
40997 |
0.19 |
| chr4_155558186_155558588 | 0.22 |
Nadk |
NAD kinase |
3991 |
0.13 |
| chr19_7523941_7524241 | 0.22 |
Atl3 |
atlastin GTPase 3 |
29556 |
0.11 |
| chr4_81508297_81508448 | 0.22 |
Gm11765 |
predicted gene 11765 |
46640 |
0.17 |
| chr4_129320984_129321142 | 0.22 |
Rbbp4 |
retinoblastoma binding protein 4, chromatin remodeling factor |
13581 |
0.11 |
| chr5_114589239_114589390 | 0.22 |
Fam222a |
family with sequence similarity 222, member A |
21297 |
0.14 |
| chr9_43545693_43545878 | 0.22 |
Gm36855 |
predicted gene, 36855 |
24781 |
0.17 |
| chr17_79658508_79658667 | 0.22 |
Rmdn2 |
regulator of microtubule dynamics 2 |
13813 |
0.21 |
| chr5_108535384_108535540 | 0.22 |
Cplx1 |
complexin 1 |
14538 |
0.11 |
| chr1_179608860_179609030 | 0.22 |
Cnst |
consortin, connexin sorting protein |
15555 |
0.18 |
| chr5_93016355_93016510 | 0.22 |
Gm25521 |
predicted gene, 25521 |
3293 |
0.18 |
| chr1_151105258_151105444 | 0.22 |
Gm19087 |
predicted gene, 19087 |
2564 |
0.2 |
| chr9_46071982_46072133 | 0.22 |
Sik3 |
SIK family kinase 3 |
51073 |
0.1 |
| chr14_30184944_30185295 | 0.21 |
Cacna1d |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
9728 |
0.2 |
| chr8_93197496_93198044 | 0.21 |
Ces1d |
carboxylesterase 1D |
7 |
0.97 |
| chr1_165470090_165470241 | 0.21 |
Gm37860 |
predicted gene, 37860 |
2665 |
0.14 |
| chr4_107388361_107388798 | 0.21 |
Ndc1 |
NDC1 transmembrane nucleoporin |
7117 |
0.15 |
| chr11_45887104_45887260 | 0.21 |
Clint1 |
clathrin interactor 1 |
4785 |
0.19 |
| chr2_166983387_166983579 | 0.21 |
Stau1 |
staufen double-stranded RNA binding protein 1 |
6712 |
0.13 |
| chr4_146575212_146575562 | 0.21 |
Gm13233 |
predicted gene 13233 |
5242 |
0.13 |
| chr11_106428672_106429050 | 0.21 |
Icam2 |
intercellular adhesion molecule 2 |
40786 |
0.1 |
| chr3_29732542_29732732 | 0.21 |
Gm37557 |
predicted gene, 37557 |
38647 |
0.19 |
| chr18_31581039_31581190 | 0.21 |
B930094E09Rik |
RIKEN cDNA B930094E09 gene |
28398 |
0.14 |
| chr16_26163874_26164039 | 0.21 |
P3h2 |
prolyl 3-hydroxylase 2 |
58172 |
0.15 |
| chr15_100110403_100110727 | 0.21 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
2404 |
0.23 |
| chr12_17885844_17886007 | 0.21 |
Gm49326 |
predicted gene, 49326 |
3728 |
0.15 |
| chr15_10637560_10637724 | 0.21 |
Gm10389 |
predicted gene 10389 |
37305 |
0.13 |
| chr13_119708466_119708617 | 0.21 |
Gm48342 |
predicted gene, 48342 |
3992 |
0.14 |
| chr12_40561876_40562268 | 0.21 |
Dock4 |
dedicator of cytokinesis 4 |
115736 |
0.06 |
| chr5_117984245_117984410 | 0.21 |
Fbxo21 |
F-box protein 21 |
5003 |
0.16 |
| chr4_109116948_109117230 | 0.21 |
Osbpl9 |
oxysterol binding protein-like 9 |
609 |
0.77 |
| chr15_78948026_78948177 | 0.21 |
Triobp |
TRIO and F-actin binding protein |
144 |
0.9 |
| chr11_58927012_58927289 | 0.21 |
Btnl10 |
butyrophilin-like 10 |
9093 |
0.06 |
| chr19_58049317_58049501 | 0.21 |
Mir5623 |
microRNA 5623 |
1758 |
0.46 |
| chr16_84806178_84806337 | 0.21 |
Jam2 |
junction adhesion molecule 2 |
3150 |
0.18 |
| chr13_67830741_67831099 | 0.21 |
4930525G20Rik |
RIKEN cDNA 4930525G20 gene |
65 |
0.66 |
| chr18_39497961_39498121 | 0.21 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
6740 |
0.28 |
| chr1_191822042_191822193 | 0.20 |
Nek2 |
NIMA (never in mitosis gene a)-related expressed kinase 2 |
65 |
0.58 |
| chr16_28841105_28841256 | 0.20 |
Mb21d2 |
Mab-21 domain containing 2 |
4813 |
0.32 |
| chr2_69423254_69423431 | 0.20 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
42897 |
0.14 |
| chr11_62409500_62409860 | 0.20 |
Ncor1 |
nuclear receptor co-repressor 1 |
8130 |
0.16 |
| chr5_24696900_24697051 | 0.20 |
Gm26648 |
predicted gene, 26648 |
7715 |
0.13 |
| chr13_98684591_98684906 | 0.20 |
Tmem171 |
transmembrane protein 171 |
10020 |
0.14 |
| chr8_40863315_40863480 | 0.20 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
819 |
0.56 |
| chr8_126920805_126921127 | 0.20 |
Gm31718 |
predicted gene, 31718 |
12751 |
0.15 |
| chr6_29540672_29541006 | 0.20 |
Irf5 |
interferon regulatory factor 5 |
307 |
0.83 |
| chr8_121561590_121561893 | 0.20 |
Fbxo31 |
F-box protein 31 |
1778 |
0.22 |
| chr17_13768386_13768604 | 0.20 |
Tcte2 |
t-complex-associated testis expressed 2 |
6670 |
0.16 |
| chr8_109972886_109973196 | 0.20 |
Gm45795 |
predicted gene 45795 |
1311 |
0.33 |
| chr1_34842105_34842416 | 0.20 |
Fam168b |
family with sequence similarity 168, member B |
704 |
0.58 |
| chr7_141581336_141581865 | 0.19 |
Ap2a2 |
adaptor-related protein complex 2, alpha 2 subunit |
19070 |
0.1 |
| chr7_73578042_73578281 | 0.19 |
1810026B05Rik |
RIKEN cDNA 1810026B05 gene |
19766 |
0.1 |
| chr2_166002813_166002989 | 0.19 |
Ncoa3 |
nuclear receptor coactivator 3 |
10264 |
0.14 |
| chr2_174499512_174499663 | 0.19 |
Gm14385 |
predicted gene 14385 |
14011 |
0.13 |
| chr15_34510659_34511041 | 0.19 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
943 |
0.43 |
| chr4_86671562_86671730 | 0.19 |
Plin2 |
perilipin 2 |
1586 |
0.38 |
| chr4_41340852_41341003 | 0.19 |
Gm26084 |
predicted gene, 26084 |
5024 |
0.12 |
| chr9_101036260_101036546 | 0.19 |
Pccb |
propionyl Coenzyme A carboxylase, beta polypeptide |
1505 |
0.33 |
| chr5_145855582_145856014 | 0.19 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
20893 |
0.14 |
| chr2_94042559_94042721 | 0.19 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
23829 |
0.14 |
| chr1_51834753_51834904 | 0.19 |
Myo1b |
myosin IB |
14282 |
0.16 |
| chr3_84011372_84011587 | 0.19 |
Tmem131l |
transmembrane 131 like |
28649 |
0.2 |
| chr10_122384642_122385270 | 0.19 |
Gm36041 |
predicted gene, 36041 |
1936 |
0.39 |
| chr13_35016682_35016867 | 0.19 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
10320 |
0.12 |
| chr11_113062597_113062759 | 0.19 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
110399 |
0.07 |
| chr1_39902031_39902182 | 0.19 |
Map4k4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
876 |
0.66 |
| chr13_4013920_4014086 | 0.19 |
Gm47853 |
predicted gene, 47853 |
9166 |
0.13 |
| chr10_112953941_112954181 | 0.19 |
Gm47519 |
predicted gene, 47519 |
6127 |
0.18 |
| chr7_123124873_123125354 | 0.19 |
Tnrc6a |
trinucleotide repeat containing 6a |
857 |
0.66 |
| chr8_116237744_116237895 | 0.18 |
4930422C21Rik |
RIKEN cDNA 4930422C21 gene |
50450 |
0.17 |
| chr8_129135025_129135198 | 0.18 |
Ccdc7b |
coiled-coil domain containing 7B |
1409 |
0.38 |
| chr5_102548279_102548672 | 0.18 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
65186 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:2001201 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.0 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.0 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid metabolic process(GO:0009698) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |