Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
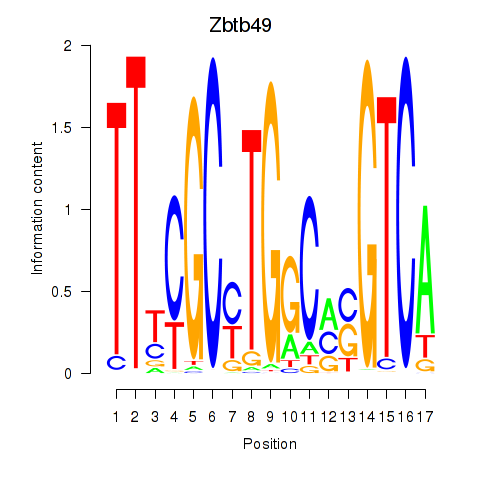
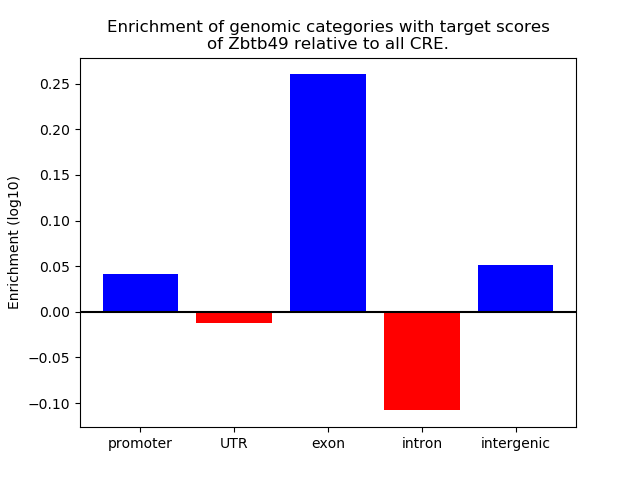
Results for Zbtb49
Z-value: 1.03
Transcription factors associated with Zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb49
|
ENSMUSG00000029127.9 | zinc finger and BTB domain containing 49 |
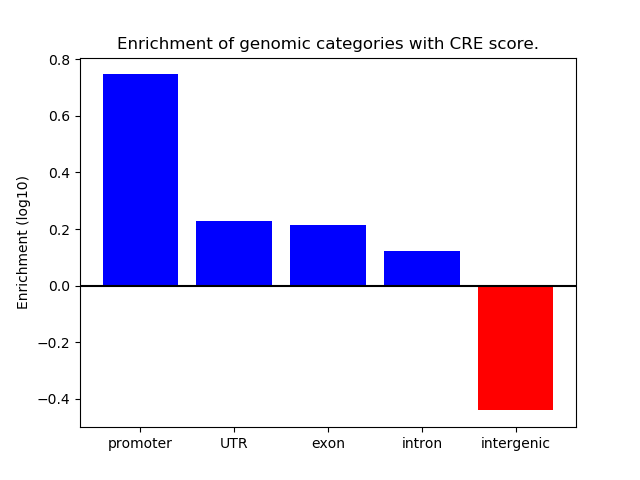
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_38220383_38220542 | Zbtb49 | 5 | 0.494346 | 0.84 | 3.6e-02 | Click! |
| chr5_38197553_38197712 | Zbtb49 | 6165 | 0.161425 | 0.47 | 3.5e-01 | Click! |
| chr5_38220127_38220304 | Zbtb49 | 198 | 0.563145 | 0.36 | 4.8e-01 | Click! |
| chr5_38219865_38220058 | Zbtb49 | 452 | 0.487467 | -0.23 | 6.6e-01 | Click! |
| chr5_38205585_38205760 | Zbtb49 | 511 | 0.743866 | -0.17 | 7.5e-01 | Click! |
Activity of the Zbtb49 motif across conditions
Conditions sorted by the z-value of the Zbtb49 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
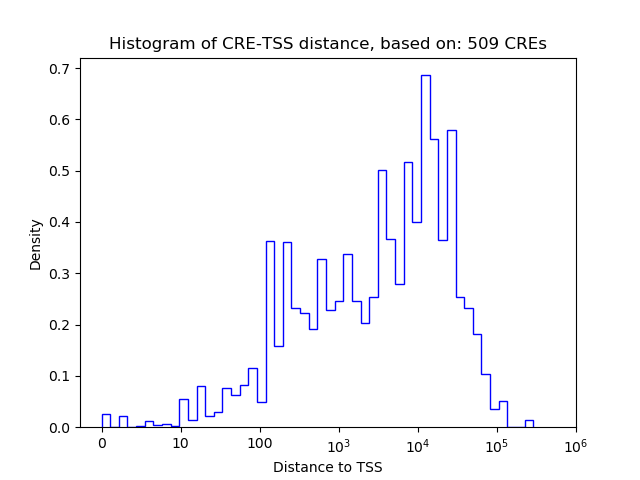
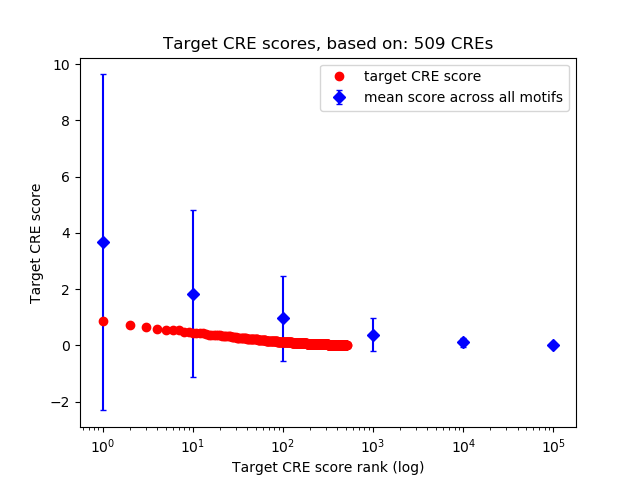
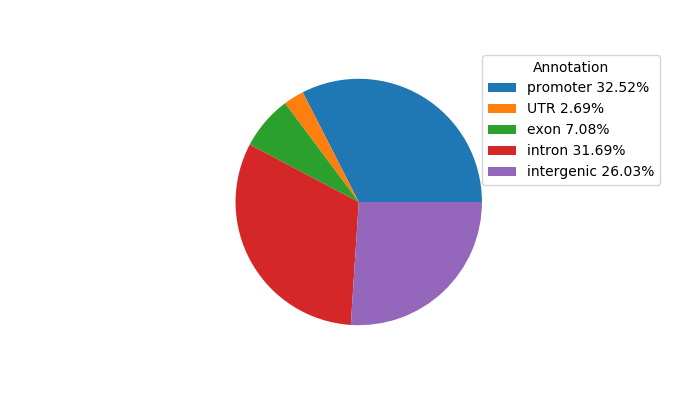
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_23964443_23964721 | 0.87 |
Bcl6 |
B cell leukemia/lymphoma 6 |
7798 |
0.17 |
| chr1_162250839_162250990 | 0.71 |
Mir214 |
microRNA 214 |
27546 |
0.14 |
| chr19_61049084_61049293 | 0.65 |
Gm22520 |
predicted gene, 22520 |
35643 |
0.14 |
| chr4_44994303_44994454 | 0.58 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
12878 |
0.1 |
| chr3_116329378_116329892 | 0.56 |
Gm29151 |
predicted gene 29151 |
20468 |
0.17 |
| chr4_49442637_49442810 | 0.54 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
8411 |
0.13 |
| chr19_39812543_39812702 | 0.54 |
Cyp2c40 |
cytochrome P450, family 2, subfamily c, polypeptide 40 |
122 |
0.97 |
| chr8_105996387_105996711 | 0.48 |
Dpep2 |
dipeptidase 2 |
126 |
0.9 |
| chr17_69417917_69418077 | 0.47 |
C030034I22Rik |
RIKEN cDNA C030034I22 gene |
536 |
0.71 |
| chr11_23124778_23124929 | 0.45 |
1700061J23Rik |
RIKEN cDNA 1700061J23 gene |
27287 |
0.14 |
| chr15_82442500_82442673 | 0.43 |
Gm49421 |
predicted gene, 49421 |
894 |
0.29 |
| chr9_32004053_32004204 | 0.43 |
Gm47465 |
predicted gene, 47465 |
1332 |
0.4 |
| chr4_148214912_148215063 | 0.43 |
Disp3 |
dispatched RND transporter family member 3 |
45764 |
0.08 |
| chr12_28898892_28899474 | 0.41 |
Gm31508 |
predicted gene, 31508 |
11046 |
0.17 |
| chr10_71234987_71235164 | 0.38 |
Tfam |
transcription factor A, mitochondrial |
246 |
0.89 |
| chr11_46085325_46085478 | 0.38 |
Mir8100 |
microRNA 8100 |
16869 |
0.12 |
| chr17_23790223_23790377 | 0.38 |
Srrm2 |
serine/arginine repetitive matrix 2 |
232 |
0.78 |
| chr3_127124044_127124345 | 0.37 |
Ank2 |
ankyrin 2, brain |
668 |
0.65 |
| chr3_94954465_94954628 | 0.36 |
Rfx5 |
regulatory factor X, 5 (influences HLA class II expression) |
19 |
0.95 |
| chr1_52454776_52454937 | 0.36 |
Gm23460 |
predicted gene, 23460 |
10312 |
0.16 |
| chr12_27357387_27357554 | 0.35 |
Sox11 |
SRY (sex determining region Y)-box 11 |
14896 |
0.28 |
| chr12_83488027_83488404 | 0.34 |
Dpf3 |
D4, zinc and double PHD fingers, family 3 |
499 |
0.78 |
| chr5_126566218_126566538 | 0.34 |
Gm24839 |
predicted gene, 24839 |
52397 |
0.14 |
| chr14_62557877_62558073 | 0.33 |
Fam124a |
family with sequence similarity 124, member A |
2200 |
0.2 |
| chr11_23855502_23855671 | 0.33 |
Papolg |
poly(A) polymerase gamma |
12884 |
0.18 |
| chr19_47304311_47304615 | 0.33 |
Sh3pxd2a |
SH3 and PX domains 2A |
10288 |
0.16 |
| chr12_111722792_111722948 | 0.30 |
Gm15995 |
predicted gene 15995 |
3522 |
0.12 |
| chr6_38531788_38531983 | 0.30 |
Fmc1 |
formation of mitochondrial complex V assembly factor 1 |
1617 |
0.3 |
| chr13_23506003_23506326 | 0.28 |
n-TStga1 |
nuclear encoded tRNA serine 1 (anticodon TGA) |
11978 |
0.05 |
| chr11_117313106_117313279 | 0.28 |
Septin9 |
septin 9 |
5038 |
0.22 |
| chr11_115859676_115859840 | 0.28 |
Myo15b |
myosin XVB |
1352 |
0.26 |
| chr4_129248394_129248565 | 0.26 |
C77080 |
expressed sequence C77080 |
36 |
0.96 |
| chr1_72231055_72231231 | 0.26 |
Gm25360 |
predicted gene, 25360 |
4903 |
0.13 |
| chr4_116008423_116008814 | 0.26 |
Faah |
fatty acid amide hydrolase |
8550 |
0.14 |
| chr17_87974584_87974753 | 0.25 |
Msh6 |
mutS homolog 6 |
394 |
0.82 |
| chr7_75861767_75861930 | 0.25 |
Klhl25 |
kelch-like 25 |
13407 |
0.22 |
| chr5_115475861_115476388 | 0.24 |
Sirt4 |
sirtuin 4 |
3814 |
0.1 |
| chr4_141708335_141708559 | 0.24 |
Ddi2 |
DNA-damage inducible protein 2 |
14972 |
0.12 |
| chr17_56467485_56467636 | 0.24 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
5344 |
0.16 |
| chr19_59588162_59588313 | 0.24 |
Gm18161 |
predicted gene, 18161 |
47786 |
0.13 |
| chr9_50856575_50856749 | 0.24 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
262 |
0.9 |
| chr4_117605417_117605587 | 0.24 |
Eri3 |
exoribonuclease 3 |
16424 |
0.15 |
| chr3_53017166_53017327 | 0.24 |
Cog6 |
component of oligomeric golgi complex 6 |
9 |
0.97 |
| chr15_75830872_75831075 | 0.22 |
Zc3h3 |
zinc finger CCCH type containing 3 |
2764 |
0.16 |
| chr12_21183631_21184095 | 0.22 |
AC156032.1 |
|
63460 |
0.08 |
| chr1_168380081_168380258 | 0.22 |
Gm38381 |
predicted gene, 38381 |
29361 |
0.18 |
| chr4_15145228_15145428 | 0.22 |
Necab1 |
N-terminal EF-hand calcium binding protein 1 |
3633 |
0.27 |
| chr2_30207072_30207249 | 0.21 |
Kyat1 |
kynurenine aminotransferase 1 |
1313 |
0.26 |
| chr4_135872055_135872460 | 0.21 |
Pnrc2 |
proline-rich nuclear receptor coactivator 2 |
804 |
0.46 |
| chr1_182336926_182337087 | 0.21 |
Fbxo28 |
F-box protein 28 |
4565 |
0.19 |
| chrX_129749825_129749976 | 0.21 |
Diaph2 |
diaphanous related formin 2 |
76 |
0.99 |
| chr19_12652059_12652547 | 0.21 |
Glyat |
glycine-N-acyltransferase |
12811 |
0.09 |
| chr10_43440139_43440316 | 0.21 |
Gm3699 |
predicted gene 3699 |
16274 |
0.13 |
| chr15_88864782_88864944 | 0.20 |
Pim3 |
proviral integration site 3 |
57 |
0.96 |
| chr3_135498924_135499245 | 0.20 |
Gm16100 |
predicted gene 16100 |
3431 |
0.15 |
| chr10_127165534_127165685 | 0.20 |
B4galnt1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
222 |
0.84 |
| chr18_64341527_64341710 | 0.19 |
Onecut2 |
one cut domain, family member 2 |
1598 |
0.35 |
| chr13_9763714_9763892 | 0.18 |
Zmynd11 |
zinc finger, MYND domain containing 11 |
547 |
0.72 |
| chr8_121575170_121575337 | 0.18 |
Fbxo31 |
F-box protein 31 |
3494 |
0.14 |
| chr11_96861486_96861931 | 0.18 |
Copz2 |
coatomer protein complex, subunit zeta 2 |
7577 |
0.09 |
| chr19_30095671_30095822 | 0.17 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
3785 |
0.25 |
| chr6_62934342_62934493 | 0.17 |
Gm5001 |
predicted gene 5001 |
5271 |
0.31 |
| chr6_108511900_108512071 | 0.17 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
3672 |
0.19 |
| chr19_33098550_33098701 | 0.17 |
Gm29946 |
predicted gene, 29946 |
24022 |
0.18 |
| chr12_116202716_116202878 | 0.17 |
Wdr60 |
WD repeat domain 60 |
11073 |
0.12 |
| chr15_36903404_36903565 | 0.17 |
Gm10384 |
predicted gene 10384 |
23668 |
0.13 |
| chr8_120409781_120409965 | 0.16 |
Gm22715 |
predicted gene, 22715 |
33676 |
0.16 |
| chr17_45592639_45593134 | 0.16 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
46 |
0.94 |
| chr7_126346739_126346890 | 0.16 |
Gm30928 |
predicted gene, 30928 |
4783 |
0.11 |
| chr17_24007509_24007674 | 0.16 |
Gm27700 |
predicted gene, 27700 |
2441 |
0.11 |
| chr11_72411311_72411561 | 0.16 |
Smtnl2 |
smoothelin-like 2 |
275 |
0.86 |
| chr14_68297827_68297985 | 0.16 |
Gm47212 |
predicted gene, 47212 |
79653 |
0.09 |
| chr11_60074031_60074200 | 0.16 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
27626 |
0.12 |
| chr19_39648534_39649207 | 0.16 |
Cyp2c67 |
cytochrome P450, family 2, subfamily c, polypeptide 67 |
181 |
0.97 |
| chr18_21216011_21216167 | 0.15 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
84034 |
0.08 |
| chr2_137115323_137115523 | 0.15 |
Jag1 |
jagged 1 |
1221 |
0.6 |
| chr4_130520077_130520248 | 0.15 |
Nkain1 |
Na+/K+ transporting ATPase interacting 1 |
37826 |
0.17 |
| chr1_187767393_187767632 | 0.15 |
AC121143.1 |
NADH dehydrogenase 2, mitochondrial (mt-Nd2) pseudogene |
54707 |
0.15 |
| chr7_14308064_14308258 | 0.14 |
Phf20-ps |
PHD finger protein 20, pseudogene |
3381 |
0.19 |
| chr5_145874134_145874475 | 0.14 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
2387 |
0.26 |
| chr15_82148269_82148913 | 0.14 |
Srebf2 |
sterol regulatory element binding factor 2 |
566 |
0.6 |
| chr8_120364518_120364684 | 0.14 |
Gm22715 |
predicted gene, 22715 |
78948 |
0.08 |
| chr18_11860532_11860692 | 0.14 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
16312 |
0.16 |
| chr12_83594536_83594711 | 0.14 |
Zfyve1 |
zinc finger, FYVE domain containing 1 |
1064 |
0.45 |
| chr2_33452045_33452383 | 0.14 |
Gm13536 |
predicted gene 13536 |
5412 |
0.16 |
| chr3_100163971_100164144 | 0.14 |
Gdap2 |
ganglioside-induced differentiation-associated-protein 2 |
1513 |
0.33 |
| chr9_86707214_86707391 | 0.13 |
Gm22138 |
predicted gene, 22138 |
3483 |
0.13 |
| chr1_59404225_59404406 | 0.13 |
Gm29016 |
predicted gene 29016 |
25385 |
0.15 |
| chr7_114292013_114292196 | 0.13 |
Psma1 |
proteasome subunit alpha 1 |
15986 |
0.21 |
| chr17_24881412_24881880 | 0.13 |
Igfals |
insulin-like growth factor binding protein, acid labile subunit |
2889 |
0.1 |
| chr5_115569868_115570025 | 0.13 |
Gcn1 |
GCN1 activator of EIF2AK4 |
4643 |
0.11 |
| chr15_78407261_78408064 | 0.13 |
Mpst |
mercaptopyruvate sulfurtransferase |
861 |
0.37 |
| chr15_35884273_35884596 | 0.13 |
Vps13b |
vacuolar protein sorting 13B |
12712 |
0.16 |
| chr2_120100029_120100210 | 0.13 |
Sptbn5 |
spectrin beta, non-erythrocytic 5 |
14441 |
0.14 |
| chr8_110716150_110716322 | 0.13 |
Mtss2 |
MTSS I-BAR domain containing 2 |
5240 |
0.18 |
| chr19_30233010_30233220 | 0.13 |
Mbl2 |
mannose-binding lectin (protein C) 2 |
173 |
0.96 |
| chr8_128466385_128467031 | 0.13 |
Nrp1 |
neuropilin 1 |
107311 |
0.06 |
| chr3_122984533_122984731 | 0.13 |
Usp53 |
ubiquitin specific peptidase 53 |
122 |
0.94 |
| chr19_32242329_32242494 | 0.12 |
Sgms1 |
sphingomyelin synthase 1 |
3585 |
0.29 |
| chr2_90504815_90505126 | 0.12 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
25720 |
0.16 |
| chr6_129254642_129254819 | 0.12 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
16005 |
0.11 |
| chr13_21945543_21945746 | 0.12 |
Zfp184 |
zinc finger protein 184 (Kruppel-like) |
149 |
0.74 |
| chr1_168475687_168476155 | 0.12 |
Mir6348 |
microRNA 6348 |
14722 |
0.24 |
| chr9_83834067_83834712 | 0.12 |
Ttk |
Ttk protein kinase |
300 |
0.91 |
| chr13_4033707_4033908 | 0.12 |
Gm18857 |
predicted gene, 18857 |
1438 |
0.31 |
| chr5_107869751_107869976 | 0.12 |
Evi5 |
ecotropic viral integration site 5 |
695 |
0.46 |
| chr11_49712994_49713225 | 0.12 |
Cnot6 |
CCR4-NOT transcription complex, subunit 6 |
386 |
0.8 |
| chr5_134295659_134295849 | 0.12 |
Gtf2i |
general transcription factor II I |
146 |
0.93 |
| chr11_23665861_23666058 | 0.12 |
Pex13 |
peroxisomal biogenesis factor 13 |
0 |
0.52 |
| chr8_71603005_71603180 | 0.12 |
Fam129c |
family with sequence similarity 129, member C |
741 |
0.42 |
| chr4_48253396_48253620 | 0.12 |
Erp44 |
endoplasmic reticulum protein 44 |
9034 |
0.2 |
| chr10_95291254_95291431 | 0.12 |
Gm48880 |
predicted gene, 48880 |
23511 |
0.13 |
| chr19_44984727_44984892 | 0.12 |
4930414N06Rik |
RIKEN cDNA 4930414N06 gene |
259 |
0.84 |
| chr10_84060915_84061106 | 0.11 |
Gm37908 |
predicted gene, 37908 |
748 |
0.67 |
| chr1_131957507_131957689 | 0.11 |
Gm29103 |
predicted gene 29103 |
5356 |
0.12 |
| chrX_20848376_20848541 | 0.11 |
Araf |
Araf proto-oncogene, serine/threonine kinase |
85 |
0.94 |
| chr11_82020261_82020449 | 0.11 |
Gm31522 |
predicted gene, 31522 |
388 |
0.79 |
| chr19_20540260_20540664 | 0.11 |
C730002L08Rik |
RIKEN cDNA C730002L08 gene |
174 |
0.96 |
| chr13_52223146_52223355 | 0.11 |
Gm48199 |
predicted gene, 48199 |
42839 |
0.19 |
| chr11_80167102_80167278 | 0.10 |
Adap2os |
ArfGAP with dual PH domains 2, opposite strand |
10439 |
0.13 |
| chr17_46646397_46646558 | 0.10 |
Mrpl2 |
mitochondrial ribosomal protein L2 |
207 |
0.69 |
| chr13_45457438_45457599 | 0.10 |
Gm47455 |
predicted gene, 47455 |
26516 |
0.17 |
| chr7_35342439_35342778 | 0.10 |
Rhpn2 |
rhophilin, Rho GTPase binding protein 2 |
8278 |
0.14 |
| chr10_77609232_77609420 | 0.10 |
Sumo3 |
small ubiquitin-like modifier 3 |
512 |
0.6 |
| chr1_105317078_105317239 | 0.10 |
Gm29014 |
predicted gene 29014 |
3963 |
0.24 |
| chr13_14630649_14630826 | 0.10 |
AW209491 |
expressed sequence AW209491 |
460 |
0.73 |
| chr5_125014623_125014796 | 0.10 |
Rflna |
refilin A |
4596 |
0.2 |
| chr15_86256321_86256487 | 0.10 |
Tbc1d22a |
TBC1 domain family, member 22a |
20899 |
0.19 |
| chr4_8693785_8693936 | 0.10 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
2495 |
0.35 |
| chr11_115832209_115832576 | 0.10 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
424 |
0.7 |
| chr19_36534453_36534632 | 0.10 |
Hectd2 |
HECT domain E3 ubiquitin protein ligase 2 |
20097 |
0.2 |
| chr2_118862147_118862313 | 0.10 |
Ivd |
isovaleryl coenzyme A dehydrogenase |
202 |
0.91 |
| chr2_120464891_120465048 | 0.09 |
Capn3 |
calpain 3 |
1007 |
0.46 |
| chr7_72247872_72248068 | 0.09 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
18610 |
0.26 |
| chr5_15549725_15549878 | 0.09 |
Gm21190 |
predicted gene, 21190 |
20579 |
0.13 |
| chr5_142768854_142769324 | 0.09 |
Tnrc18 |
trinucleotide repeat containing 18 |
3067 |
0.26 |
| chr7_131257681_131257832 | 0.09 |
Cdcp3 |
CUB domain containing protein 3 |
47503 |
0.09 |
| chr11_85183469_85183675 | 0.09 |
1700125H20Rik |
RIKEN cDNA 1700125H20 gene |
12476 |
0.13 |
| chr2_25126889_25127229 | 0.09 |
AL732309.1 |
exonuclease 3'-5' domain containing 3 (EXD3) pseudogene |
10778 |
0.08 |
| chr16_92428960_92429111 | 0.09 |
Gm46555 |
predicted gene, 46555 |
28442 |
0.11 |
| chr16_93729531_93729931 | 0.09 |
Dop1b |
DOP1 leucine zipper like protein B |
368 |
0.85 |
| chr7_110096322_110096530 | 0.09 |
Zfp143 |
zinc finger protein 143 |
4797 |
0.14 |
| chr10_4334368_4334532 | 0.09 |
Akap12 |
A kinase (PRKA) anchor protein (gravin) 12 |
95 |
0.97 |
| chr19_6291962_6292220 | 0.09 |
Ehd1 |
EH-domain containing 1 |
2090 |
0.13 |
| chr7_39818171_39818337 | 0.09 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
20512 |
0.15 |
| chr17_47926111_47926270 | 0.09 |
Foxp4 |
forkhead box P4 |
1545 |
0.29 |
| chr14_73325714_73325873 | 0.09 |
Rb1 |
RB transcriptional corepressor 1 |
29 |
0.98 |
| chr7_99230091_99230271 | 0.09 |
Mogat2 |
monoacylglycerol O-acyltransferase 2 |
8413 |
0.12 |
| chr7_128461344_128461499 | 0.09 |
Tial1 |
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
66 |
0.9 |
| chr15_55547224_55547377 | 0.08 |
Mrpl13 |
mitochondrial ribosomal protein L13 |
864 |
0.61 |
| chr17_83935315_83935481 | 0.08 |
Gm23905 |
predicted gene, 23905 |
12190 |
0.11 |
| chr8_111053951_111054114 | 0.08 |
Exosc6 |
exosome component 6 |
2307 |
0.15 |
| chr14_16598201_16598430 | 0.08 |
Rarb |
retinoic acid receptor, beta |
22488 |
0.21 |
| chr12_71768786_71769132 | 0.08 |
Gm47556 |
predicted gene, 47556 |
959 |
0.59 |
| chr10_8133541_8133713 | 0.08 |
Gm30906 |
predicted gene, 30906 |
42504 |
0.18 |
| chr7_102313337_102313504 | 0.08 |
Stim1 |
stromal interaction molecule 1 |
45093 |
0.09 |
| chr9_48605290_48605498 | 0.08 |
Nnmt |
nicotinamide N-methyltransferase |
241 |
0.95 |
| chr9_13297643_13297990 | 0.08 |
Maml2 |
mastermind like transcriptional coactivator 2 |
141 |
0.95 |
| chr4_155356437_155356594 | 0.08 |
Prkcz |
protein kinase C, zeta |
4825 |
0.16 |
| chr8_88199324_88199484 | 0.08 |
Tent4b |
terminal nucleotidyltransferase 4B |
190 |
0.93 |
| chr13_41226373_41226543 | 0.08 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
6053 |
0.13 |
| chr19_21031007_21031192 | 0.08 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
73403 |
0.1 |
| chr6_121865645_121865799 | 0.08 |
Mug1 |
murinoglobulin 1 |
19855 |
0.18 |
| chr8_84980308_84980512 | 0.08 |
Junb |
jun B proto-oncogene |
1692 |
0.13 |
| chr18_76241500_76241662 | 0.08 |
Smad2 |
SMAD family member 2 |
1 |
0.98 |
| chr11_96788704_96788867 | 0.08 |
Cbx1 |
chromobox 1 |
342 |
0.79 |
| chr11_5402470_5402635 | 0.07 |
Znrf3 |
zinc and ring finger 3 |
20812 |
0.18 |
| chr5_70735708_70735881 | 0.07 |
Gabrg1 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
106410 |
0.07 |
| chr5_53469821_53470044 | 0.07 |
Rbpj |
recombination signal binding protein for immunoglobulin kappa J region |
3780 |
0.25 |
| chr11_96035249_96035421 | 0.07 |
Snf8 |
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
370 |
0.72 |
| chr4_119233398_119233632 | 0.07 |
P3h1 |
prolyl 3-hydroxylase 1 |
285 |
0.7 |
| chr14_21522234_21522697 | 0.07 |
Kat6b |
K(lysine) acetyltransferase 6B |
5590 |
0.25 |
| chr8_87093816_87094174 | 0.07 |
Gm5118 |
predicted gene 5118 |
688 |
0.66 |
| chr14_34298539_34299045 | 0.07 |
Shld2 |
shieldin complex subunit 2 |
1537 |
0.24 |
| chr5_38561684_38561885 | 0.07 |
Wdr1 |
WD repeat domain 1 |
22 |
0.97 |
| chr19_30241056_30241233 | 0.07 |
Mbl2 |
mannose-binding lectin (protein C) 2 |
8202 |
0.21 |
| chr3_108911697_108911858 | 0.07 |
Prpf38b |
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
50 |
0.96 |
| chr8_45794944_45795095 | 0.07 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
8189 |
0.16 |
| chr6_108773223_108773381 | 0.07 |
Arl8b |
ADP-ribosylation factor-like 8B |
9797 |
0.23 |
| chr9_65607205_65607476 | 0.07 |
Pif1 |
PIF1 5'-to-3' DNA helicase |
20114 |
0.13 |
| chr17_83525682_83525983 | 0.07 |
Cox7a2l |
cytochrome c oxidase subunit 7A2 like |
8502 |
0.26 |
| chr7_53604334_53604505 | 0.07 |
Mir6238 |
microRNA 6238 |
287470 |
0.01 |
| chr2_28840575_28840979 | 0.07 |
Ddx31 |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 31 |
318 |
0.56 |
| chr13_93442726_93443017 | 0.06 |
Gm6109 |
predicted gene 6109 |
5617 |
0.16 |
| chr11_120831406_120831995 | 0.06 |
Gm11773 |
predicted gene 11773 |
671 |
0.52 |
| chr1_75010536_75010846 | 0.06 |
Nhej1 |
non-homologous end joining factor 1 |
35948 |
0.1 |
| chr13_58756508_58756704 | 0.06 |
Rpl13-ps2 |
ribosomal protein L13, pseudogene 2 |
55 |
0.96 |
| chr2_131881708_131882048 | 0.06 |
Erv3 |
endogenous retroviral sequence 3 |
22131 |
0.1 |
| chr10_42277646_42277818 | 0.06 |
Foxo3 |
forkhead box O3 |
977 |
0.64 |
| chr10_62651277_62651445 | 0.06 |
Ddx50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
143 |
0.93 |
| chr16_29944225_29944722 | 0.06 |
Gm26569 |
predicted gene, 26569 |
2043 |
0.31 |
| chr16_77501096_77501279 | 0.06 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
803 |
0.51 |
| chr5_130266836_130266987 | 0.06 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
7981 |
0.13 |
| chr15_10719115_10719326 | 0.06 |
4930556M19Rik |
RIKEN cDNA 4930556M19 gene |
1369 |
0.44 |
| chr5_136171356_136171939 | 0.06 |
Orai2 |
ORAI calcium release-activated calcium modulator 2 |
934 |
0.4 |
| chr1_40169151_40169486 | 0.06 |
Il1r1 |
interleukin 1 receptor, type I |
55762 |
0.1 |
| chr9_48339748_48339930 | 0.06 |
Nxpe2 |
neurexophilin and PC-esterase domain family, member 2 |
995 |
0.58 |
| chr4_126262494_126262652 | 0.06 |
Trappc3 |
trafficking protein particle complex 3 |
173 |
0.92 |
| chr5_32311758_32312085 | 0.06 |
Gm15615 |
predicted gene 15615 |
198 |
0.94 |
| chr8_111538280_111538723 | 0.06 |
Znrf1 |
zinc and ring finger 1 |
36 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.0 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |