Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
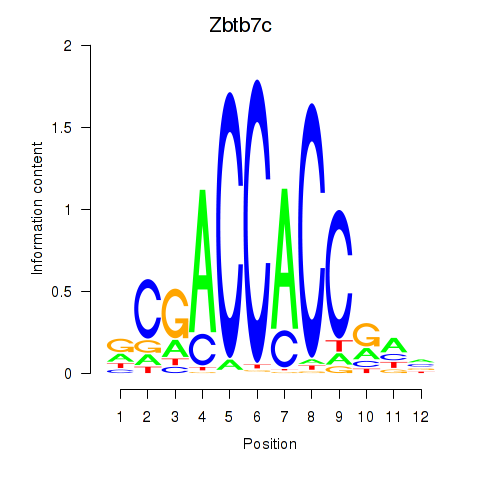
Results for Zbtb7c
Z-value: 1.36
Transcription factors associated with Zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7c
|
ENSMUSG00000044646.8 | zinc finger and BTB domain containing 7C |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
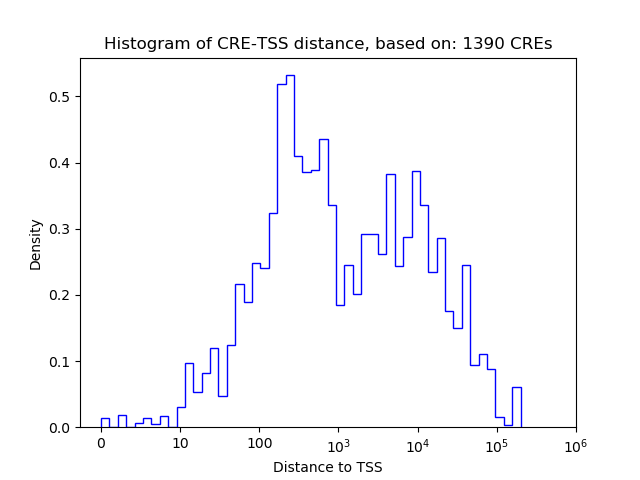
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_75901792_75902127 | Zbtb7c | 81744 | 0.090063 | 0.99 | 6.3e-05 | Click! |
| chr18_75817816_75817971 | Zbtb7c | 2285 | 0.341032 | -0.97 | 1.3e-03 | Click! |
| chr18_75902133_75902303 | Zbtb7c | 82003 | 0.089691 | 0.96 | 2.6e-03 | Click! |
| chr18_75837745_75837910 | Zbtb7c | 17612 | 0.220943 | 0.80 | 5.6e-02 | Click! |
| chr18_76077358_76077509 | Zbtb7c | 17975 | 0.186045 | -0.77 | 7.2e-02 | Click! |
Activity of the Zbtb7c motif across conditions
Conditions sorted by the z-value of the Zbtb7c motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
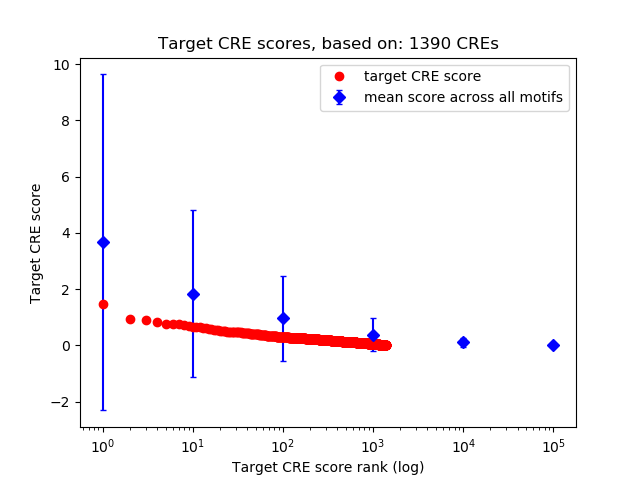
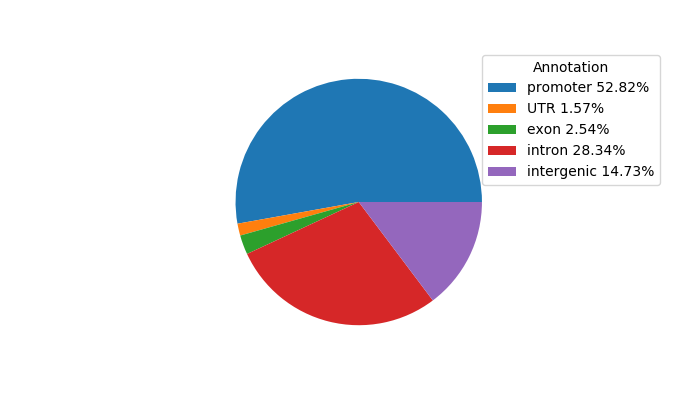
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_75173089_75173918 | 1.48 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr4_33310456_33310613 | 0.92 |
Rngtt |
RNA guanylyltransferase and 5'-phosphatase |
206 |
0.7 |
| chr9_67840208_67840482 | 0.92 |
Vps13c |
vacuolar protein sorting 13C |
51 |
0.98 |
| chr18_46280937_46281101 | 0.84 |
Pggt1b |
protein geranylgeranyltransferase type I, beta subunit |
26 |
0.97 |
| chr4_53123043_53123194 | 0.77 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
36777 |
0.16 |
| chrY_90839675_90839839 | 0.76 |
Gm21748 |
predicted gene, 21748 |
580 |
0.73 |
| chr18_61423942_61424123 | 0.75 |
Gm8755 |
predicted gene 8755 |
10169 |
0.12 |
| chrY_90755303_90755481 | 0.71 |
Gm21860 |
predicted gene, 21860 |
75 |
0.97 |
| chr16_90199492_90199754 | 0.69 |
Gm49704 |
predicted gene, 49704 |
1475 |
0.35 |
| chr3_156999583_156999772 | 0.67 |
Gm43527 |
predicted gene 43527 |
740 |
0.74 |
| chr11_96075628_96076164 | 0.66 |
Atp5g1 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
226 |
0.86 |
| chr13_54323124_54323298 | 0.64 |
Gm48622 |
predicted gene, 48622 |
4669 |
0.18 |
| chr9_48984872_48985421 | 0.62 |
Usp28 |
ubiquitin specific peptidase 28 |
229 |
0.92 |
| chr5_129941480_129942081 | 0.62 |
Vkorc1l1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
190 |
0.89 |
| chr8_85555040_85555462 | 0.57 |
Dnaja2 |
DnaJ heat shock protein family (Hsp40) member A2 |
93 |
0.97 |
| chr6_117907668_117908056 | 0.57 |
4933440N22Rik |
RIKEN cDNA 4933440N22 gene |
102 |
0.91 |
| chr8_28274310_28274461 | 0.56 |
Gm8100 |
predicted gene 8100 |
88545 |
0.09 |
| chr10_80343063_80343448 | 0.55 |
Adamtsl5 |
ADAMTS-like 5 |
1983 |
0.12 |
| chr1_67160568_67161028 | 0.55 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
37772 |
0.17 |
| chr14_68809096_68809247 | 0.50 |
Gm47256 |
predicted gene, 47256 |
65664 |
0.11 |
| chr4_8763954_8764148 | 0.50 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
12613 |
0.28 |
| chr18_60743844_60744030 | 0.49 |
Rps14 |
ribosomal protein S14 |
3161 |
0.19 |
| chr13_37600818_37601107 | 0.49 |
Gm47753 |
predicted gene, 47753 |
18691 |
0.1 |
| chr10_75780213_75780364 | 0.49 |
Gm20441 |
predicted gene 20441 |
666 |
0.36 |
| chr3_129744894_129745074 | 0.49 |
Gm42650 |
predicted gene 42650 |
356 |
0.81 |
| chr9_98986714_98987086 | 0.48 |
Faim |
Fas apoptotic inhibitory molecule |
493 |
0.73 |
| chr3_108383463_108383620 | 0.47 |
Gm12522 |
predicted gene 12522 |
200 |
0.59 |
| chr16_25879136_25879302 | 0.46 |
Gm4524 |
predicted gene 4524 |
77256 |
0.1 |
| chr12_30911832_30911990 | 0.46 |
Sh3yl1 |
Sh3 domain YSC-like 1 |
219 |
0.62 |
| chr6_35538904_35539680 | 0.46 |
Mtpn |
myotrophin |
507 |
0.82 |
| chr16_38097146_38097299 | 0.46 |
Gsk3b |
glycogen synthase kinase 3 beta |
6934 |
0.23 |
| chr18_82668033_82668343 | 0.46 |
Zfp236 |
zinc finger protein 236 |
15380 |
0.1 |
| chr14_74735013_74735164 | 0.46 |
Esd |
esterase D/formylglutathione hydrolase |
567 |
0.78 |
| chr2_32524318_32524490 | 0.46 |
Gm13412 |
predicted gene 13412 |
627 |
0.55 |
| chr14_61680870_61681697 | 0.45 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr17_81376267_81376581 | 0.45 |
Gm50044 |
predicted gene, 50044 |
5591 |
0.28 |
| chr9_108346931_108347091 | 0.44 |
Usp4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
842 |
0.34 |
| chr8_13889986_13890137 | 0.44 |
Coprs |
coordinator of PRMT5, differentiation stimulator |
71 |
0.95 |
| chr5_149635750_149636168 | 0.44 |
Hsph1 |
heat shock 105kDa/110kDa protein 1 |
212 |
0.91 |
| chr18_11230345_11230496 | 0.43 |
Gata6 |
GATA binding protein 6 |
171373 |
0.03 |
| chr10_80577672_80578424 | 0.43 |
Klf16 |
Kruppel-like factor 16 |
727 |
0.41 |
| chr19_57453287_57453451 | 0.42 |
Trub1 |
TruB pseudouridine (psi) synthase family member 1 |
412 |
0.82 |
| chr2_167330209_167330360 | 0.42 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
16384 |
0.17 |
| chr11_118724289_118724499 | 0.42 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
36647 |
0.17 |
| chr12_56345567_56345749 | 0.42 |
Mbip |
MAP3K12 binding inhibitory protein 1 |
163 |
0.95 |
| chr2_30270553_30270704 | 0.41 |
Phyhd1 |
phytanoyl-CoA dioxygenase domain containing 1 |
3765 |
0.11 |
| chr8_46492911_46493695 | 0.41 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
471 |
0.78 |
| chr10_81600790_81601143 | 0.41 |
Tle6 |
transducin-like enhancer of split 6 |
66 |
0.93 |
| chr10_82699476_82699690 | 0.41 |
Hcfc2 |
host cell factor C2 |
372 |
0.82 |
| chr7_127026907_127027653 | 0.40 |
Maz |
MYC-associated zinc finger protein (purine-binding transcription factor) |
243 |
0.69 |
| chr8_72135457_72135608 | 0.40 |
Tpm4 |
tropomyosin 4 |
274 |
0.79 |
| chr4_40144303_40144840 | 0.40 |
Aco1 |
aconitase 1 |
1490 |
0.41 |
| chr15_99044356_99044570 | 0.39 |
Prph |
peripherin |
10711 |
0.08 |
| chr12_112140078_112140229 | 0.39 |
Kif26a |
kinesin family member 26A |
6055 |
0.14 |
| chr6_90469663_90469814 | 0.38 |
Klf15 |
Kruppel-like factor 15 |
2620 |
0.15 |
| chr4_105169027_105169380 | 0.38 |
Plpp3 |
phospholipid phosphatase 3 |
11856 |
0.27 |
| chr1_74067073_74067224 | 0.37 |
Tns1 |
tensin 1 |
24203 |
0.17 |
| chr16_37565521_37565674 | 0.37 |
Rabl3 |
RAB, member RAS oncogene family-like 3 |
1863 |
0.27 |
| chr17_88621894_88622400 | 0.37 |
Ston1 |
stonin 1 |
4408 |
0.19 |
| chr11_73048007_73048735 | 0.37 |
Ncbp3 |
nuclear cap binding subunit 3 |
498 |
0.7 |
| chr7_90476395_90476725 | 0.37 |
Dlg2 |
discs large MAGUK scaffold protein 2 |
112 |
0.86 |
| chr10_53378945_53379463 | 0.36 |
Gm47641 |
predicted gene, 47641 |
248 |
0.79 |
| chr9_78192346_78192497 | 0.36 |
Gsta4 |
glutathione S-transferase, alpha 4 |
474 |
0.69 |
| chr2_163299510_163299661 | 0.36 |
Tox2 |
TOX high mobility group box family member 2 |
20793 |
0.19 |
| chr7_34647419_34647585 | 0.36 |
Kctd15 |
potassium channel tetramerisation domain containing 15 |
1242 |
0.37 |
| chr16_17200424_17201140 | 0.35 |
Ube2l3 |
ubiquitin-conjugating enzyme E2L 3 |
677 |
0.49 |
| chr2_155062362_155062698 | 0.35 |
Gm45609 |
predicted gene 45609 |
11651 |
0.13 |
| chr8_20550460_20550749 | 0.34 |
Gm21182 |
predicted gene, 21182 |
6882 |
0.19 |
| chr2_180589607_180589819 | 0.34 |
Ogfr |
opioid growth factor receptor |
251 |
0.89 |
| chr5_100518822_100518973 | 0.34 |
Cops4 |
COP9 signalosome subunit 4 |
160 |
0.9 |
| chr9_58253443_58253626 | 0.34 |
Stoml1 |
stomatin-like 1 |
342 |
0.81 |
| chr9_55544376_55544543 | 0.34 |
Isl2 |
insulin related protein 2 (islet 2) |
2123 |
0.24 |
| chr5_33722581_33722796 | 0.33 |
Fgfr3 |
fibroblast growth factor receptor 3 |
132 |
0.9 |
| chr8_114439821_114440145 | 0.33 |
Wwox |
WW domain-containing oxidoreductase |
281 |
0.92 |
| chr12_87157684_87158165 | 0.33 |
Gstz1 |
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
316 |
0.8 |
| chr13_64031193_64031358 | 0.33 |
Hsd17b3 |
hydroxysteroid (17-beta) dehydrogenase 3 |
31858 |
0.15 |
| chr7_127604746_127604924 | 0.32 |
1700120K04Rik |
RIKEN cDNA 1700120K04 gene |
390 |
0.65 |
| chr14_65837485_65837656 | 0.32 |
Ccdc25 |
coiled-coil domain containing 25 |
210 |
0.93 |
| chr3_95906804_95907175 | 0.32 |
Car14 |
carbonic anhydrase 14 |
2298 |
0.13 |
| chr3_121953604_121953967 | 0.32 |
Arhgap29 |
Rho GTPase activating protein 29 |
555 |
0.78 |
| chr6_29507708_29507870 | 0.32 |
Gm26627 |
predicted gene, 26627 |
64 |
0.61 |
| chr9_57710332_57710483 | 0.31 |
Edc3 |
enhancer of mRNA decapping 3 |
1840 |
0.24 |
| chr4_61969528_61969679 | 0.31 |
Mup-ps19 |
major urinary protein, pseudogene 19 |
9692 |
0.17 |
| chr7_114066489_114066748 | 0.31 |
Gm45615 |
predicted gene 45615 |
20280 |
0.22 |
| chr11_94165237_94165388 | 0.31 |
B230206L02Rik |
RIKEN cDNA B230206L02 gene |
29652 |
0.15 |
| chr17_29773464_29773650 | 0.31 |
Gm50016 |
predicted gene, 50016 |
45830 |
0.09 |
| chr1_31018175_31018326 | 0.31 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
18346 |
0.14 |
| chr2_163301297_163301733 | 0.31 |
Tox2 |
TOX high mobility group box family member 2 |
18863 |
0.19 |
| chr19_53902427_53902597 | 0.31 |
Pdcd4 |
programmed cell death 4 |
787 |
0.57 |
| chr2_31504934_31505459 | 0.30 |
Ass1 |
argininosuccinate synthetase 1 |
4470 |
0.21 |
| chr2_154435738_154435889 | 0.30 |
Cbfa2t2 |
CBFA2/RUNX1 translocation partner 2 |
668 |
0.68 |
| chr2_181366266_181366439 | 0.30 |
Zgpat |
zinc finger, CCCH-type with G patch domain |
900 |
0.29 |
| chr17_46111130_46111298 | 0.30 |
Mrps18a |
mitochondrial ribosomal protein S18A |
218 |
0.89 |
| chr7_99199162_99199329 | 0.30 |
Gm45012 |
predicted gene 45012 |
3121 |
0.17 |
| chr12_80135777_80135928 | 0.30 |
2310015A10Rik |
RIKEN cDNA 2310015A10 gene |
3008 |
0.18 |
| chr8_61535951_61536102 | 0.30 |
Palld |
palladin, cytoskeletal associated protein |
2185 |
0.38 |
| chr2_166714253_166714603 | 0.29 |
Prex1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
596 |
0.76 |
| chr17_71255942_71256093 | 0.29 |
Emilin2 |
elastin microfibril interfacer 2 |
329 |
0.86 |
| chr4_118234344_118235000 | 0.29 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
1344 |
0.4 |
| chr10_16717618_16717784 | 0.29 |
Gm5948 |
predicted gene 5948 |
51239 |
0.14 |
| chr6_51681649_51681815 | 0.29 |
Gm38811 |
predicted gene, 38811 |
29349 |
0.19 |
| chr4_45981155_45981352 | 0.29 |
Tdrd7 |
tudor domain containing 7 |
8419 |
0.21 |
| chr16_5146310_5146979 | 0.29 |
Sec14l5 |
SEC14-like lipid binding 5 |
465 |
0.73 |
| chr15_59076755_59077073 | 0.29 |
Mtss1 |
MTSS I-BAR domain containing 1 |
5075 |
0.26 |
| chr1_91463744_91463928 | 0.29 |
Per2 |
period circadian clock 2 |
4512 |
0.13 |
| chr11_117115576_117115749 | 0.29 |
Sec14l1 |
SEC14-like lipid binding 1 |
410 |
0.75 |
| chr2_134843502_134843690 | 0.29 |
Gm14036 |
predicted gene 14036 |
39647 |
0.17 |
| chr19_17775291_17775442 | 0.28 |
Pcsk5 |
proprotein convertase subtilisin/kexin type 5 |
62251 |
0.11 |
| chrX_134058988_134059612 | 0.28 |
Cstf2 |
cleavage stimulation factor, 3' pre-RNA subunit 2 |
11 |
0.97 |
| chr6_17463758_17464495 | 0.28 |
Met |
met proto-oncogene |
19 |
0.98 |
| chr4_123749751_123749929 | 0.28 |
Akirin1 |
akirin 1 |
391 |
0.81 |
| chr9_106202795_106203252 | 0.28 |
Twf2 |
twinfilin actin binding protein 2 |
85 |
0.93 |
| chr4_116827683_116827841 | 0.28 |
Gm12996 |
predicted gene 12996 |
2859 |
0.12 |
| chr16_36875280_36875431 | 0.28 |
Golgb1 |
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
153 |
0.48 |
| chr17_79893522_79893693 | 0.28 |
Atl2 |
atlastin GTPase 2 |
2444 |
0.27 |
| chr19_8373854_8374041 | 0.28 |
Slc22a30 |
solute carrier family 22, member 30 |
31113 |
0.16 |
| chr10_80674829_80675669 | 0.28 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
1044 |
0.31 |
| chr11_120549054_120549419 | 0.28 |
Mcrip1 |
MAPK regulated corepressor interacting protein 1 |
465 |
0.54 |
| chrX_75972880_75973041 | 0.27 |
Gm7155 |
predicted gene 7155 |
163 |
0.95 |
| chr14_118001652_118001971 | 0.27 |
Dct |
dopachrome tautomerase |
25724 |
0.18 |
| chr6_51533320_51533471 | 0.27 |
Snx10 |
sorting nexin 10 |
9110 |
0.21 |
| chr5_114690986_114691154 | 0.27 |
Gltp |
glycolipid transfer protein |
86 |
0.95 |
| chr2_35049177_35049382 | 0.27 |
Hc |
hemolytic complement |
12159 |
0.16 |
| chr7_118544510_118544696 | 0.27 |
Coq7 |
demethyl-Q 7 |
11247 |
0.14 |
| chr6_141876497_141876758 | 0.27 |
Gm30784 |
predicted gene, 30784 |
10269 |
0.2 |
| chr2_51145877_51146125 | 0.27 |
Rnd3 |
Rho family GTPase 3 |
3093 |
0.32 |
| chr15_59044955_59045265 | 0.27 |
Mtss1 |
MTSS I-BAR domain containing 1 |
4513 |
0.25 |
| chr14_66153104_66153314 | 0.27 |
Ptk2b |
PTK2 protein tyrosine kinase 2 beta |
7469 |
0.17 |
| chr12_8674756_8674907 | 0.27 |
Pum2 |
pumilio RNA-binding family member 2 |
66 |
0.98 |
| chr5_24428552_24428786 | 0.27 |
Slc4a2 |
solute carrier family 4 (anion exchanger), member 2 |
246 |
0.79 |
| chr14_26669361_26669540 | 0.27 |
Mir7672 |
microRNA 7672 |
158 |
0.69 |
| chr3_14868267_14868499 | 0.27 |
Car3 |
carbonic anhydrase 3 |
795 |
0.63 |
| chr13_34874226_34874546 | 0.27 |
Prpf4b |
pre-mRNA processing factor 4B |
916 |
0.34 |
| chr13_14035661_14035873 | 0.26 |
Tbce |
tubulin-specific chaperone E |
3812 |
0.14 |
| chrX_37126911_37127100 | 0.26 |
Nkap |
NFKB activating protein |
210 |
0.89 |
| chr7_27838642_27838925 | 0.26 |
Zfp59 |
zinc finger protein 59 |
176 |
0.92 |
| chr4_116450069_116450227 | 0.26 |
Gm12950 |
predicted gene 12950 |
1360 |
0.39 |
| chr19_10882123_10882346 | 0.26 |
Tmem109 |
transmembrane protein 109 |
233 |
0.86 |
| chr10_89624588_89625080 | 0.26 |
Slc17a8 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
3581 |
0.24 |
| chr19_42504942_42505277 | 0.26 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
13650 |
0.19 |
| chr9_74884159_74884330 | 0.26 |
Onecut1 |
one cut domain, family member 1 |
17760 |
0.15 |
| chr8_106893764_106894131 | 0.26 |
Utp4 |
UTP4 small subunit processome component |
296 |
0.38 |
| chrX_159255089_159255806 | 0.26 |
Rps6ka3 |
ribosomal protein S6 kinase polypeptide 3 |
335 |
0.92 |
| chr3_157906196_157906370 | 0.26 |
Cth |
cystathionase (cystathionine gamma-lyase) |
558 |
0.68 |
| chr12_77501624_77501785 | 0.26 |
Gm48177 |
predicted gene, 48177 |
24694 |
0.17 |
| chr7_98360800_98361039 | 0.26 |
Tsku |
tsukushi, small leucine rich proteoglycan |
369 |
0.85 |
| chr5_125478134_125478423 | 0.26 |
Gm27551 |
predicted gene, 27551 |
1099 |
0.36 |
| chr19_55118166_55118344 | 0.26 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
8948 |
0.2 |
| chr10_79996620_79996838 | 0.26 |
Abca7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
235 |
0.81 |
| chr7_99176016_99176394 | 0.26 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
4088 |
0.15 |
| chr9_69332142_69332305 | 0.26 |
Gm15511 |
predicted gene 15511 |
32024 |
0.15 |
| chr2_160731435_160731699 | 0.26 |
Plcg1 |
phospholipase C, gamma 1 |
132 |
0.96 |
| chr6_38353981_38354166 | 0.26 |
Zc3hav1 |
zinc finger CCCH type, antiviral 1 |
200 |
0.91 |
| chr16_76454192_76454374 | 0.25 |
Gm45030 |
predicted gene 45030 |
48626 |
0.13 |
| chr10_80057662_80057905 | 0.25 |
Gpx4 |
glutathione peroxidase 4 |
1785 |
0.18 |
| chrX_12761746_12762400 | 0.25 |
Med14 |
mediator complex subunit 14 |
0 |
0.73 |
| chr5_74401642_74401799 | 0.25 |
Scfd2 |
Sec1 family domain containing 2 |
3982 |
0.2 |
| chr11_116306188_116306606 | 0.25 |
Exoc7 |
exocyst complex component 7 |
255 |
0.84 |
| chr4_105204876_105205027 | 0.25 |
Plpp3 |
phospholipid phosphatase 3 |
47604 |
0.17 |
| chr11_72796331_72796482 | 0.25 |
Zzef1 |
zinc finger, ZZ-type with EF hand domain 1 |
152 |
0.65 |
| chr1_63112731_63113827 | 0.25 |
Ino80dos |
INO80 complex subunit D, opposite strand |
201 |
0.84 |
| chr9_78587729_78587880 | 0.25 |
Slc17a5 |
solute carrier family 17 (anion/sugar transporter), member 5 |
211 |
0.92 |
| chr11_60454740_60455113 | 0.25 |
Drg2 |
developmentally regulated GTP binding protein 2 |
302 |
0.81 |
| chr2_90928885_90929161 | 0.25 |
Ptpmt1 |
protein tyrosine phosphatase, mitochondrial 1 |
10765 |
0.1 |
| chr5_123022889_123023101 | 0.25 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
7657 |
0.09 |
| chr19_23142022_23142190 | 0.25 |
Klf9 |
Kruppel-like factor 9 |
880 |
0.54 |
| chr14_20733586_20734177 | 0.25 |
Ndst2 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
681 |
0.49 |
| chr17_31389981_31390174 | 0.25 |
Pde9a |
phosphodiesterase 9A |
3783 |
0.18 |
| chr2_170130193_170130373 | 0.25 |
Zfp217 |
zinc finger protein 217 |
937 |
0.7 |
| chr13_48966249_48966400 | 0.25 |
Fam120a |
family with sequence similarity 120, member A |
1693 |
0.44 |
| chr6_88037325_88037512 | 0.24 |
Gm44187 |
predicted gene, 44187 |
5049 |
0.12 |
| chr9_87021835_87022144 | 0.24 |
Cyb5r4 |
cytochrome b5 reductase 4 |
25 |
0.97 |
| chr3_92080492_92080974 | 0.24 |
Lor |
loricrin |
2409 |
0.17 |
| chr5_134455761_134455912 | 0.24 |
Gtf2ird1 |
general transcription factor II I repeat domain-containing 1 |
115 |
0.54 |
| chr12_98716508_98716741 | 0.24 |
Ptpn21 |
protein tyrosine phosphatase, non-receptor type 21 |
17189 |
0.12 |
| chr16_93311764_93312102 | 0.24 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
41256 |
0.14 |
| chr15_36794017_36794623 | 0.24 |
Ywhaz |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
211 |
0.92 |
| chr19_44554722_44555277 | 0.24 |
Ndufb8 |
NADH:ubiquinone oxidoreductase subunit B8 |
13 |
0.96 |
| chr6_118478873_118479106 | 0.24 |
Zfp9 |
zinc finger protein 9 |
331 |
0.85 |
| chr2_167347223_167347603 | 0.24 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
1770 |
0.36 |
| chr9_109875655_109876424 | 0.24 |
Cdc25a |
cell division cycle 25A |
99 |
0.94 |
| chr6_116064944_116065095 | 0.24 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
6970 |
0.18 |
| chr1_88049643_88049948 | 0.24 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
5593 |
0.08 |
| chr13_75730593_75731025 | 0.24 |
Gm48302 |
predicted gene, 48302 |
3530 |
0.17 |
| chr11_50324830_50325400 | 0.24 |
Canx |
calnexin |
558 |
0.65 |
| chr17_62656805_62657225 | 0.24 |
Gm25800 |
predicted gene, 25800 |
199895 |
0.03 |
| chr9_61269634_61269800 | 0.24 |
B930092H01Rik |
RIKEN cDNA B930092H01 gene |
24092 |
0.18 |
| chr19_55101157_55101316 | 0.24 |
Gm31356 |
predicted gene, 31356 |
1734 |
0.32 |
| chr8_95142522_95142681 | 0.24 |
Kifc3 |
kinesin family member C3 |
54 |
0.96 |
| chr5_23412404_23412614 | 0.24 |
Gm42509 |
predicted gene 42509 |
16406 |
0.11 |
| chr12_84317174_84317418 | 0.24 |
Zfp410 |
zinc finger protein 410 |
151 |
0.58 |
| chr2_67898259_67898744 | 0.24 |
Gm37964 |
predicted gene, 37964 |
95 |
0.98 |
| chr11_116624217_116624394 | 0.24 |
Rhbdf2 |
rhomboid 5 homolog 2 |
58 |
0.95 |
| chr19_6987629_6987792 | 0.23 |
Vegfb |
vascular endothelial growth factor B |
59 |
0.92 |
| chr11_78262090_78262390 | 0.23 |
2610507B11Rik |
RIKEN cDNA 2610507B11 gene |
232 |
0.82 |
| chr3_51159499_51159691 | 0.23 |
Gm38246 |
predicted gene, 38246 |
55935 |
0.09 |
| chr17_27204481_27205047 | 0.23 |
Lemd2 |
LEM domain containing 2 |
295 |
0.81 |
| chr9_122128202_122128353 | 0.23 |
4632418H02Rik |
RIKEN cDNA 4632418H02 gene |
983 |
0.42 |
| chr5_125385398_125385549 | 0.23 |
Ubc |
ubiquitin C |
3713 |
0.14 |
| chr12_75488348_75488526 | 0.23 |
Gm47690 |
predicted gene, 47690 |
20308 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.2 | 0.9 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 1.7 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 0.4 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.3 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.2 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.1 | 0.2 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.7 | GO:0039535 | regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.4 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 0.3 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.2 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.1 | 0.2 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.1 | 0.3 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.3 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.4 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.0 | 0.3 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.0 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.0 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.0 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.0 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:2000758 | positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:1902996 | regulation of tau-protein kinase activity(GO:1902947) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.0 | GO:0006057 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 1.0 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0018599 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.1 | 0.3 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0034784 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.4 | GO:0044688 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0018651 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.8 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |