Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
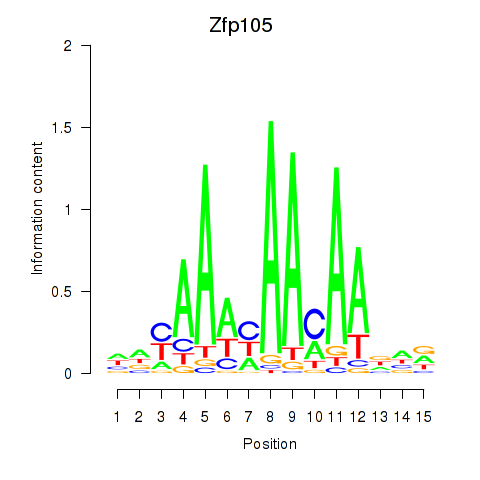
Results for Zfp105
Z-value: 1.92
Transcription factors associated with Zfp105
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp105
|
ENSMUSG00000057895.5 | zinc finger protein 105 |
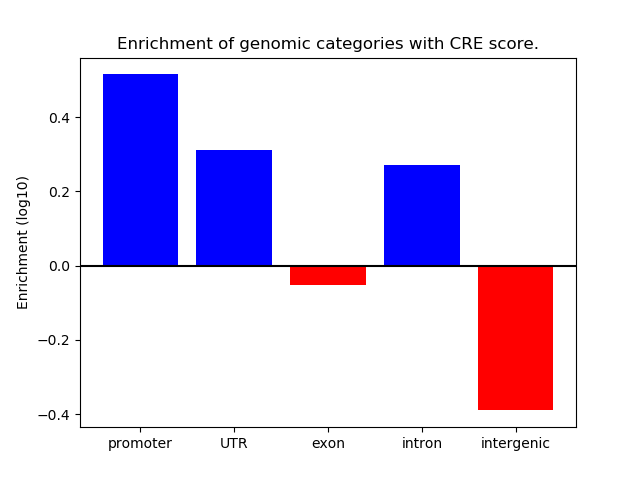
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_122922994_122923156 | Zfp105 | 3 | 0.944647 | 0.04 | 9.5e-01 | Click! |
| chr9_122923241_122923469 | Zfp105 | 256 | 0.815973 | 0.03 | 9.6e-01 | Click! |
Activity of the Zfp105 motif across conditions
Conditions sorted by the z-value of the Zfp105 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
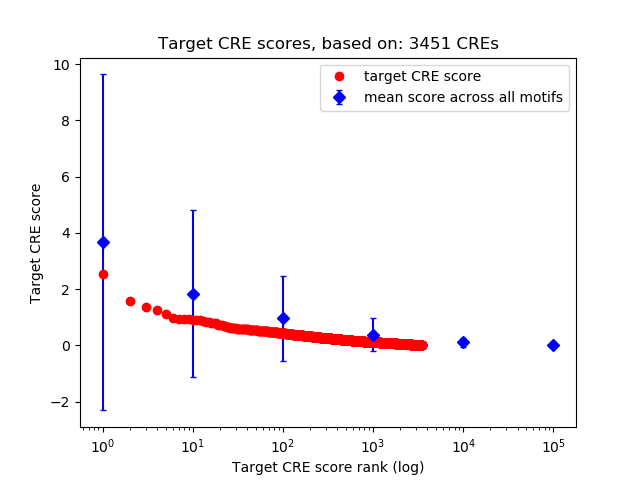
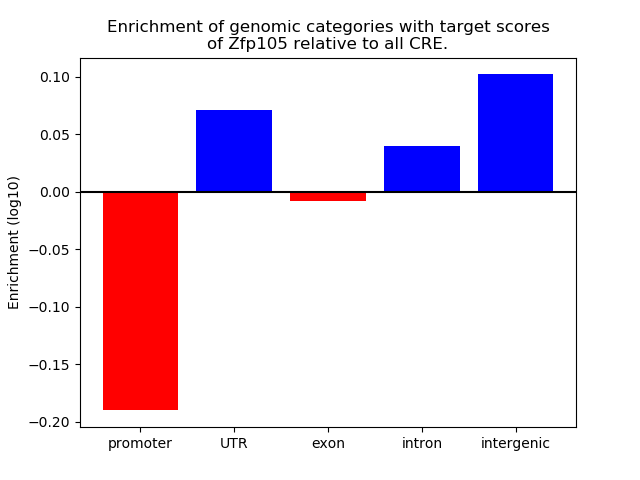
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_46137654_46137874 | 2.52 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
475 |
0.71 |
| chr10_59786684_59786837 | 1.57 |
Gm17059 |
predicted gene 17059 |
13494 |
0.14 |
| chr9_45391079_45391243 | 1.38 |
Fxyd2 |
FXYD domain-containing ion transport regulator 2 |
8508 |
0.12 |
| chr2_103849595_103849746 | 1.27 |
Gm13879 |
predicted gene 13879 |
6014 |
0.09 |
| chr10_8086566_8086745 | 1.10 |
Gm48614 |
predicted gene, 48614 |
65363 |
0.12 |
| chr1_79486426_79486688 | 0.98 |
Gm19027 |
predicted gene, 19027 |
20600 |
0.19 |
| chr19_46138051_46138212 | 0.94 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
842 |
0.48 |
| chr12_40596811_40597378 | 0.93 |
Dock4 |
dedicator of cytokinesis 4 |
150758 |
0.04 |
| chr7_44810745_44811033 | 0.93 |
Atf5 |
activating transcription factor 5 |
4769 |
0.08 |
| chr5_117096808_117097327 | 0.91 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
1744 |
0.29 |
| chr6_121889143_121889294 | 0.91 |
Mug1 |
murinoglobulin 1 |
3641 |
0.23 |
| chr12_21183631_21184095 | 0.90 |
AC156032.1 |
|
63460 |
0.08 |
| chr8_70221071_70221246 | 0.85 |
Armc6 |
armadillo repeat containing 6 |
1132 |
0.32 |
| chr11_57983626_57984029 | 0.83 |
Gm12249 |
predicted gene 12249 |
10693 |
0.15 |
| chr4_138342269_138342429 | 0.82 |
Cda |
cytidine deaminase |
1300 |
0.3 |
| chr9_20443751_20443910 | 0.80 |
Zfp26 |
zinc finger protein 26 |
2456 |
0.16 |
| chr2_103822101_103822252 | 0.79 |
Gm13880 |
predicted gene 13880 |
2700 |
0.13 |
| chr7_123193136_123193287 | 0.79 |
Tnrc6a |
trinucleotide repeat containing 6a |
13290 |
0.2 |
| chr11_117266799_117266976 | 0.73 |
Septin9 |
septin 9 |
641 |
0.7 |
| chr6_97871999_97872401 | 0.73 |
Gm15531 |
predicted gene 15531 |
18339 |
0.23 |
| chr10_61313715_61314040 | 0.73 |
Rpl27a-ps1 |
ribosomal protein L27A, pseudogene 1 |
4047 |
0.16 |
| chr4_141679306_141679614 | 0.69 |
Rsc1a1 |
regulatory solute carrier protein, family 1, member 1 |
6139 |
0.13 |
| chr19_30128744_30128991 | 0.67 |
Gldc |
glycine decarboxylase |
16364 |
0.18 |
| chr13_45891782_45891969 | 0.66 |
Atxn1 |
ataxin 1 |
19587 |
0.21 |
| chr10_20285755_20285929 | 0.65 |
Gm48249 |
predicted gene, 48249 |
784 |
0.5 |
| chr6_50175016_50175173 | 0.63 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
5206 |
0.25 |
| chr3_101881841_101882004 | 0.62 |
Slc22a15 |
solute carrier family 22 (organic anion/cation transporter), member 15 |
4035 |
0.26 |
| chr2_52923177_52923340 | 0.62 |
Fmnl2 |
formin-like 2 |
65390 |
0.13 |
| chr3_93600843_93601099 | 0.61 |
Gm5541 |
predicted gene 5541 |
28781 |
0.09 |
| chr2_103626307_103626554 | 0.61 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
60120 |
0.1 |
| chr13_98595769_98595925 | 0.60 |
Gm4815 |
predicted gene 4815 |
17654 |
0.12 |
| chr9_53596608_53596772 | 0.59 |
Acat1 |
acetyl-Coenzyme A acetyltransferase 1 |
12933 |
0.14 |
| chr7_109704650_109704808 | 0.59 |
Akip1 |
A kinase (PRKA) interacting protein 1 |
977 |
0.32 |
| chr1_168476281_168476457 | 0.59 |
Mir6348 |
microRNA 6348 |
14274 |
0.25 |
| chr13_114853110_114853434 | 0.59 |
Mocs2 |
molybdenum cofactor synthesis 2 |
34367 |
0.17 |
| chr13_93630762_93631092 | 0.58 |
Gm15622 |
predicted gene 15622 |
5545 |
0.17 |
| chr7_113265601_113265755 | 0.57 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
25924 |
0.15 |
| chr11_70712647_70712798 | 0.57 |
Mir6925 |
microRNA 6925 |
6732 |
0.08 |
| chr15_31117050_31117219 | 0.57 |
Gm26416 |
predicted gene, 26416 |
77204 |
0.08 |
| chr10_83327701_83327852 | 0.57 |
Slc41a2 |
solute carrier family 41, member 2 |
4893 |
0.18 |
| chr7_80559132_80559287 | 0.57 |
Blm |
Bloom syndrome, RecQ like helicase |
24090 |
0.13 |
| chr16_13190073_13190384 | 0.56 |
Ercc4 |
excision repair cross-complementing rodent repair deficiency, complementation group 4 |
57837 |
0.13 |
| chr17_31858140_31858307 | 0.56 |
Sik1 |
salt inducible kinase 1 |
2419 |
0.24 |
| chr2_102745048_102745199 | 0.56 |
Gm13872 |
predicted gene 13872 |
1623 |
0.43 |
| chr11_93364300_93364451 | 0.55 |
Gm24856 |
predicted gene, 24856 |
48949 |
0.19 |
| chr9_77757821_77758009 | 0.55 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
3380 |
0.18 |
| chr4_141124895_141125054 | 0.54 |
4921514A10Rik |
RIKEN cDNA 4921514A10 gene |
9314 |
0.11 |
| chr12_118281460_118281611 | 0.54 |
Sp4 |
trans-acting transcription factor 4 |
19833 |
0.22 |
| chr15_36471981_36472313 | 0.53 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr15_88865691_88865931 | 0.53 |
Pim3 |
proviral integration site 3 |
891 |
0.49 |
| chr11_59972001_59972167 | 0.53 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
574 |
0.65 |
| chr18_46614972_46615447 | 0.53 |
Gm3734 |
predicted gene 3734 |
15598 |
0.13 |
| chr11_106913695_106913846 | 0.53 |
Smurf2 |
SMAD specific E3 ubiquitin protein ligase 2 |
6505 |
0.15 |
| chr6_99522653_99522811 | 0.52 |
Foxp1 |
forkhead box P1 |
11 |
0.98 |
| chr10_8228115_8228580 | 0.52 |
Gm30906 |
predicted gene, 30906 |
52216 |
0.15 |
| chr4_149794929_149795189 | 0.52 |
Gm13065 |
predicted gene 13065 |
1640 |
0.21 |
| chr12_8217539_8217881 | 0.52 |
Gm33037 |
predicted gene, 33037 |
8545 |
0.19 |
| chr2_126584960_126585111 | 0.51 |
Hdc |
histidine decarboxylase |
13226 |
0.17 |
| chr18_23496939_23497187 | 0.51 |
Dtna |
dystrobrevin alpha |
228 |
0.96 |
| chr15_102437859_102438043 | 0.51 |
Amhr2 |
anti-Mullerian hormone type 2 receptor |
7416 |
0.1 |
| chr6_145767466_145768203 | 0.50 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
17263 |
0.18 |
| chr2_103870054_103870389 | 0.50 |
Gm13876 |
predicted gene 13876 |
18103 |
0.09 |
| chr12_21144740_21144943 | 0.50 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
32887 |
0.16 |
| chr4_132234389_132234540 | 0.50 |
Gmeb1 |
glucocorticoid modulatory element binding protein 1 |
442 |
0.62 |
| chr1_62423315_62423491 | 0.50 |
Pard3bos3 |
par-3 family cell polarity regulator beta, opposite strand 3 |
75858 |
0.1 |
| chr9_112743581_112744166 | 0.50 |
Gm24957 |
predicted gene, 24957 |
221344 |
0.02 |
| chr6_125040762_125040928 | 0.49 |
Ing4 |
inhibitor of growth family, member 4 |
984 |
0.24 |
| chr13_93625562_93625714 | 0.49 |
Gm15622 |
predicted gene 15622 |
256 |
0.91 |
| chr13_45903603_45903795 | 0.49 |
Atxn1 |
ataxin 1 |
31411 |
0.17 |
| chr12_113252720_113252871 | 0.48 |
Gm25622 |
predicted gene, 25622 |
38646 |
0.1 |
| chr11_106578107_106578307 | 0.48 |
Tex2 |
testis expressed gene 2 |
906 |
0.59 |
| chr9_110657562_110657931 | 0.48 |
Ccdc12 |
coiled-coil domain containing 12 |
1134 |
0.29 |
| chr14_55080666_55080850 | 0.48 |
Zfhx2 |
zinc finger homeobox 2 |
306 |
0.77 |
| chr17_63494033_63494531 | 0.47 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
1529 |
0.49 |
| chr1_165625442_165625599 | 0.47 |
Mpzl1 |
myelin protein zero-like 1 |
8566 |
0.12 |
| chr8_21810141_21810292 | 0.47 |
Gm15316 |
predicted gene 15316 |
7168 |
0.1 |
| chr18_20976494_20976645 | 0.47 |
Rnf125 |
ring finger protein 125 |
15097 |
0.19 |
| chr17_46809254_46809414 | 0.47 |
Bicral |
BRD4 interacting chromatin remodeling complex associated protein like |
2851 |
0.18 |
| chr18_46708132_46708283 | 0.46 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
19822 |
0.12 |
| chr9_77845751_77846338 | 0.46 |
Gm19572 |
predicted gene, 19572 |
5664 |
0.15 |
| chr11_49088297_49088883 | 0.46 |
Gm12188 |
predicted gene 12188 |
47 |
0.79 |
| chr11_119946799_119947314 | 0.46 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
3584 |
0.14 |
| chr16_24223745_24224269 | 0.46 |
Gm31814 |
predicted gene, 31814 |
7483 |
0.23 |
| chr15_99711244_99711472 | 0.46 |
Smarcd1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
3478 |
0.09 |
| chr4_26470101_26470279 | 0.46 |
Gm11903 |
predicted gene 11903 |
27729 |
0.19 |
| chr1_179836199_179836367 | 0.46 |
Ahctf1 |
AT hook containing transcription factor 1 |
32603 |
0.16 |
| chr9_65526608_65526759 | 0.46 |
Gm17749 |
predicted gene, 17749 |
3123 |
0.17 |
| chr2_41790211_41790364 | 0.46 |
Lrp1b |
low density lipoprotein-related protein 1B |
1209 |
0.66 |
| chr5_87135400_87135653 | 0.45 |
Ugt2b5 |
UDP glucuronosyltransferase 2 family, polypeptide B5 |
4792 |
0.14 |
| chr19_46915492_46915672 | 0.45 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
6007 |
0.19 |
| chr5_142648566_142648753 | 0.44 |
Wipi2 |
WD repeat domain, phosphoinositide interacting 2 |
13855 |
0.15 |
| chr5_144234168_144234324 | 0.44 |
Bri3 |
brain protein I3 |
10191 |
0.11 |
| chr10_13866673_13866990 | 0.44 |
Aig1 |
androgen-induced 1 |
1991 |
0.24 |
| chr1_79815049_79815281 | 0.44 |
Serpine2 |
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
5139 |
0.21 |
| chr1_93142666_93142824 | 0.44 |
Agxt |
alanine-glyoxylate aminotransferase |
2866 |
0.18 |
| chr2_69199630_69199898 | 0.44 |
Spc25 |
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
5366 |
0.17 |
| chr6_93879123_93879274 | 0.44 |
Gm22840 |
predicted gene, 22840 |
11417 |
0.24 |
| chr17_32250617_32250775 | 0.44 |
Gm4432 |
predicted gene 4432 |
8533 |
0.13 |
| chr15_102655668_102655956 | 0.44 |
Gm4544 |
predicted gene 4544 |
1299 |
0.28 |
| chr12_18606247_18606408 | 0.43 |
Gm48398 |
predicted gene, 48398 |
4057 |
0.19 |
| chr11_32138403_32138577 | 0.43 |
Gm12108 |
predicted gene 12108 |
37262 |
0.11 |
| chr10_84859659_84859818 | 0.43 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
21590 |
0.19 |
| chr11_102282845_102283012 | 0.43 |
Tmub2 |
transmembrane and ubiquitin-like domain containing 2 |
2003 |
0.18 |
| chr15_80741946_80742097 | 0.43 |
Gm49512 |
predicted gene, 49512 |
29750 |
0.13 |
| chr6_114785531_114785684 | 0.42 |
Gm44331 |
predicted gene, 44331 |
3783 |
0.21 |
| chr14_40939013_40939164 | 0.42 |
Tspan14 |
tetraspanin 14 |
4767 |
0.23 |
| chr1_189923142_189923294 | 0.42 |
Smyd2 |
SET and MYND domain containing 2 |
855 |
0.59 |
| chrX_12240420_12240590 | 0.42 |
Gm14521 |
predicted gene 14521 |
60438 |
0.13 |
| chr9_53596297_53596463 | 0.42 |
Acat1 |
acetyl-Coenzyme A acetyltransferase 1 |
12623 |
0.14 |
| chr2_92178970_92179191 | 0.41 |
Phf21a |
PHD finger protein 21A |
5026 |
0.19 |
| chr15_36472644_36473037 | 0.41 |
Ankrd46 |
ankyrin repeat domain 46 |
23875 |
0.13 |
| chr18_8766236_8766411 | 0.41 |
Gm26119 |
predicted gene, 26119 |
67967 |
0.11 |
| chr6_5447010_5447161 | 0.41 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
49176 |
0.15 |
| chrX_12253274_12253448 | 0.40 |
Gm14521 |
predicted gene 14521 |
73294 |
0.1 |
| chr13_102925968_102926119 | 0.40 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
19937 |
0.27 |
| chr3_94399233_94399394 | 0.40 |
Lingo4 |
leucine rich repeat and Ig domain containing 4 |
796 |
0.33 |
| chr10_59442892_59443079 | 0.40 |
Oit3 |
oncoprotein induced transcript 3 |
1207 |
0.45 |
| chr8_33800813_33800977 | 0.40 |
Rbpms |
RNA binding protein gene with multiple splicing |
3681 |
0.19 |
| chr5_120495968_120496124 | 0.40 |
Plbd2 |
phospholipase B domain containing 2 |
7555 |
0.11 |
| chr11_88643557_88643735 | 0.39 |
Msi2 |
musashi RNA-binding protein 2 |
53499 |
0.13 |
| chr13_34725685_34725960 | 0.39 |
Gm47151 |
predicted gene, 47151 |
5071 |
0.13 |
| chr8_3402101_3402252 | 0.39 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
9103 |
0.17 |
| chr5_151086750_151086901 | 0.39 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
2437 |
0.36 |
| chr8_45283600_45283787 | 0.39 |
Klkb1 |
kallikrein B, plasma 1 |
11166 |
0.15 |
| chr8_41214926_41215280 | 0.39 |
Fgl1 |
fibrinogen-like protein 1 |
50 |
0.97 |
| chr2_157550429_157550705 | 0.39 |
Gm14286 |
predicted gene 14286 |
5098 |
0.12 |
| chr9_44127033_44127184 | 0.39 |
Mcam |
melanoma cell adhesion molecule |
7361 |
0.06 |
| chr8_106572917_106573101 | 0.39 |
Gm10073 |
predicted pseudogene 10073 |
341 |
0.86 |
| chr8_107696511_107696668 | 0.39 |
Gm8940 |
predicted gene 8940 |
5745 |
0.27 |
| chr7_112515425_112515576 | 0.38 |
Parva |
parvin, alpha |
4201 |
0.25 |
| chr16_77434703_77434862 | 0.38 |
9430053O09Rik |
RIKEN cDNA 9430053O09 gene |
12962 |
0.11 |
| chr8_101353164_101353334 | 0.38 |
Gm22223 |
predicted gene, 22223 |
186459 |
0.03 |
| chr19_41812412_41812564 | 0.38 |
Arhgap19 |
Rho GTPase activating protein 19 |
10441 |
0.15 |
| chr11_3267258_3267426 | 0.38 |
Drg1 |
developmentally regulated GTP binding protein 1 |
927 |
0.45 |
| chr3_131474115_131474266 | 0.38 |
Sgms2 |
sphingomyelin synthase 2 |
16289 |
0.23 |
| chr13_11455361_11455526 | 0.38 |
Gm25496 |
predicted gene, 25496 |
28545 |
0.21 |
| chr3_66410712_66410863 | 0.38 |
Gm50014 |
predicted gene, 50014 |
46248 |
0.17 |
| chr12_78248706_78249176 | 0.38 |
Gm18899 |
predicted gene, 18899 |
7494 |
0.15 |
| chr6_121861209_121861360 | 0.38 |
Mug1 |
murinoglobulin 1 |
20180 |
0.18 |
| chr15_36827079_36827245 | 0.37 |
Gm49282 |
predicted gene, 49282 |
15654 |
0.14 |
| chr12_44947878_44948344 | 0.37 |
Gm15902 |
predicted gene 15902 |
16782 |
0.21 |
| chr1_170640120_170640365 | 0.37 |
Olfml2b |
olfactomedin-like 2B |
4290 |
0.21 |
| chr6_128103977_128104136 | 0.37 |
Gm26338 |
predicted gene, 26338 |
9030 |
0.15 |
| chr11_4521480_4521785 | 0.37 |
Mtmr3 |
myotubularin related protein 3 |
6774 |
0.2 |
| chr8_14825690_14825955 | 0.37 |
Dlgap2 |
DLG associated protein 2 |
48049 |
0.12 |
| chr11_23858437_23858588 | 0.37 |
Papolg |
poly(A) polymerase gamma |
9958 |
0.19 |
| chr4_44992680_44993053 | 0.37 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
11366 |
0.1 |
| chr11_94390845_94391254 | 0.37 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
1871 |
0.29 |
| chr6_128523521_128524414 | 0.37 |
Pzp |
PZP, alpha-2-macroglobulin like |
2736 |
0.13 |
| chr12_102437143_102437359 | 0.37 |
Lgmn |
legumain |
2562 |
0.25 |
| chr4_102734911_102735096 | 0.36 |
Gm24869 |
predicted gene, 24869 |
1809 |
0.4 |
| chr3_96541676_96541827 | 0.36 |
Gm15441 |
predicted gene 15441 |
16064 |
0.07 |
| chr10_63413324_63413532 | 0.36 |
Gm7530 |
predicted gene 7530 |
429 |
0.77 |
| chr2_68125158_68125473 | 0.36 |
B3galt1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
7602 |
0.26 |
| chr19_36626660_36626979 | 0.36 |
Hectd2os |
Hectd2, opposite strand |
795 |
0.68 |
| chr18_11829207_11829416 | 0.36 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
9909 |
0.16 |
| chr5_8902050_8902207 | 0.36 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
8179 |
0.15 |
| chr9_111211265_111211429 | 0.36 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
2846 |
0.24 |
| chr5_145857663_145857814 | 0.36 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
18953 |
0.15 |
| chr10_77062455_77062639 | 0.36 |
Col18a1 |
collagen, type XVIII, alpha 1 |
2550 |
0.22 |
| chr17_35486151_35486302 | 0.36 |
H2-Q10 |
histocompatibility 2, Q region locus 10 |
16129 |
0.07 |
| chr10_56500437_56500624 | 0.35 |
Gm9118 |
predicted gene 9118 |
3189 |
0.31 |
| chr4_116562433_116562592 | 0.35 |
Gpbp1l1 |
GC-rich promoter binding protein 1-like 1 |
4433 |
0.12 |
| chr19_34521505_34521678 | 0.35 |
Lipa |
lysosomal acid lipase A |
5820 |
0.14 |
| chr5_121200253_121200404 | 0.35 |
Rpl6 |
ribosomal protein L6 |
4153 |
0.14 |
| chr8_17664465_17664717 | 0.35 |
Csmd1 |
CUB and Sushi multiple domains 1 |
129005 |
0.06 |
| chr5_149171514_149171679 | 0.35 |
Gm43332 |
predicted gene 43332 |
11587 |
0.08 |
| chrX_164035471_164035631 | 0.35 |
Car5b |
carbonic anhydrase 5b, mitochondrial |
7554 |
0.2 |
| chr10_61486909_61487060 | 0.35 |
Gm47595 |
predicted gene, 47595 |
9306 |
0.1 |
| chr10_51721782_51721985 | 0.35 |
Rfx6 |
regulatory factor X, 6 |
28112 |
0.12 |
| chr13_43232894_43233157 | 0.34 |
Tbc1d7 |
TBC1 domain family, member 7 |
61524 |
0.1 |
| chr10_77898806_77898957 | 0.34 |
Lrrc3 |
leucine rich repeat containing 3 |
3655 |
0.09 |
| chr6_145360391_145360595 | 0.34 |
Gm23498 |
predicted gene, 23498 |
6365 |
0.15 |
| chr12_109991265_109991416 | 0.34 |
Gm34667 |
predicted gene, 34667 |
32533 |
0.1 |
| chr16_8532772_8532923 | 0.34 |
Abat |
4-aminobutyrate aminotransferase |
19362 |
0.15 |
| chr1_174771619_174771985 | 0.34 |
Fmn2 |
formin 2 |
57476 |
0.15 |
| chr8_10946099_10946520 | 0.34 |
Gm44955 |
predicted gene 44955 |
1581 |
0.25 |
| chr7_12774535_12774865 | 0.34 |
Zscan18 |
zinc finger and SCAN domain containing 18 |
361 |
0.71 |
| chr15_76575802_76576193 | 0.34 |
Adck5 |
aarF domain containing kinase 5 |
361 |
0.67 |
| chr11_102211007_102211317 | 0.34 |
Hdac5 |
histone deacetylase 5 |
7766 |
0.09 |
| chr2_173974366_173974517 | 0.33 |
Atp5k-ps2 |
ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E, pseudogene 2 |
10250 |
0.2 |
| chr5_9045319_9045483 | 0.33 |
Gm40264 |
predicted gene, 40264 |
10277 |
0.15 |
| chr6_87866715_87866872 | 0.33 |
Gm44064 |
predicted gene, 44064 |
15584 |
0.08 |
| chr3_17862499_17862757 | 0.33 |
Gm23441 |
predicted gene, 23441 |
58881 |
0.1 |
| chr7_97622705_97622904 | 0.33 |
Rsf1os1 |
remodeling and spacing factor 1, opposite strand 1 |
22308 |
0.1 |
| chr2_102751919_102752079 | 0.33 |
Slc1a2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
3908 |
0.25 |
| chr8_88983561_88983712 | 0.33 |
Mir8110 |
microRNA 8110 |
41099 |
0.17 |
| chr4_118398583_118398989 | 0.33 |
Szt2 |
SZT2 subunit of KICSTOR complex |
8853 |
0.11 |
| chr1_14795269_14795483 | 0.33 |
Gm37412 |
predicted gene, 37412 |
6731 |
0.2 |
| chr11_68656972_68657153 | 0.33 |
Myh10 |
myosin, heavy polypeptide 10, non-muscle |
34497 |
0.15 |
| chr2_167681702_167681863 | 0.33 |
Cebpb |
CCAAT/enhancer binding protein (C/EBP), beta |
7133 |
0.11 |
| chr15_62039645_62039829 | 0.33 |
Pvt1 |
Pvt1 oncogene |
480 |
0.86 |
| chr5_121407443_121407594 | 0.32 |
Gm43420 |
predicted gene 43420 |
4860 |
0.11 |
| chr18_46737499_46737680 | 0.32 |
Atg12 |
autophagy related 12 |
3968 |
0.14 |
| chr10_12858109_12858285 | 0.32 |
Utrn |
utrophin |
3538 |
0.24 |
| chr7_35131018_35131347 | 0.32 |
Gm35665 |
predicted gene, 35665 |
10267 |
0.1 |
| chr2_32692363_32692549 | 0.32 |
Fpgs |
folylpolyglutamyl synthetase |
788 |
0.35 |
| chr10_7954404_7954895 | 0.32 |
Tab2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
1244 |
0.52 |
| chr6_147064387_147064568 | 0.32 |
Mrps35 |
mitochondrial ribosomal protein S35 |
3226 |
0.17 |
| chr6_5509398_5509797 | 0.32 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
13288 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.3 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 2.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.2 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.2 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:2001201 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.2 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.1 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0071351 | cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.0 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.2 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0000237 | leptotene(GO:0000237) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.2 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.0 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.0 | 0.0 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.2 | GO:0017014 | protein nitrosylation(GO:0017014) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.1 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.0 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.0 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.0 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) establishment of T cell polarity(GO:0001768) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.0 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.0 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0043830 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.0 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |