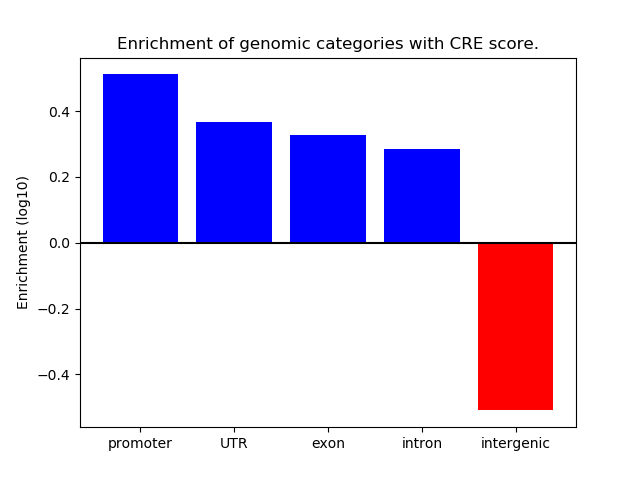
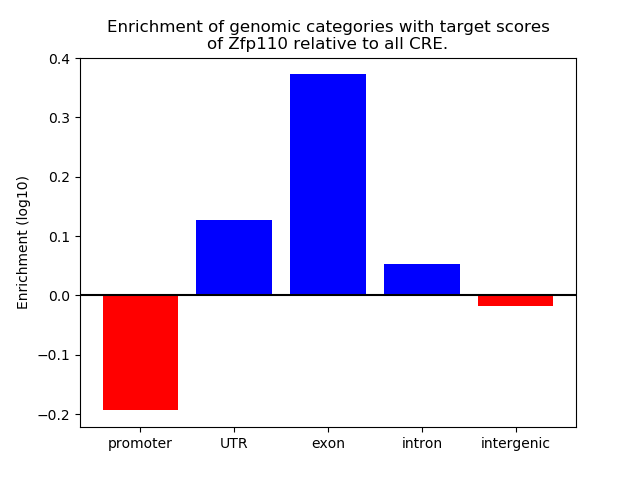
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
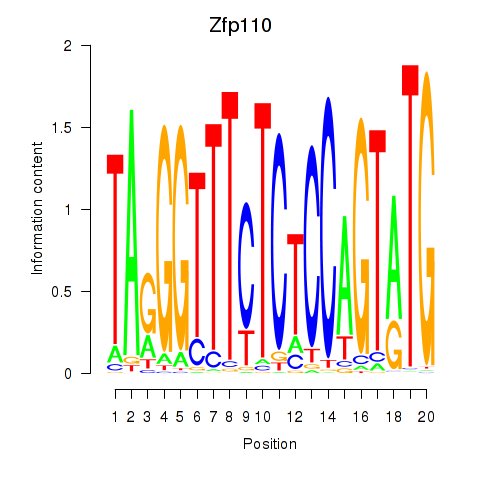
Results for Zfp110
Z-value: 0.86
Transcription factors associated with Zfp110
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp110
|
ENSMUSG00000058638.7 | zinc finger protein 110 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_12834899_12835063 | Zfp110 | 56 | 0.935136 | -0.94 | 4.7e-03 | Click! |
| chr7_12834640_12834802 | Zfp110 | 40 | 0.938700 | -0.90 | 1.5e-02 | Click! |
| chr7_12834378_12834558 | Zfp110 | 293 | 0.776086 | 0.44 | 3.8e-01 | Click! |
| chr7_12835149_12835365 | Zfp110 | 220 | 0.840543 | 0.22 | 6.7e-01 | Click! |
| chr7_12835382_12835707 | Zfp110 | 61 | 0.933905 | 0.16 | 7.6e-01 | Click! |
Activity of the Zfp110 motif across conditions
Conditions sorted by the z-value of the Zfp110 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
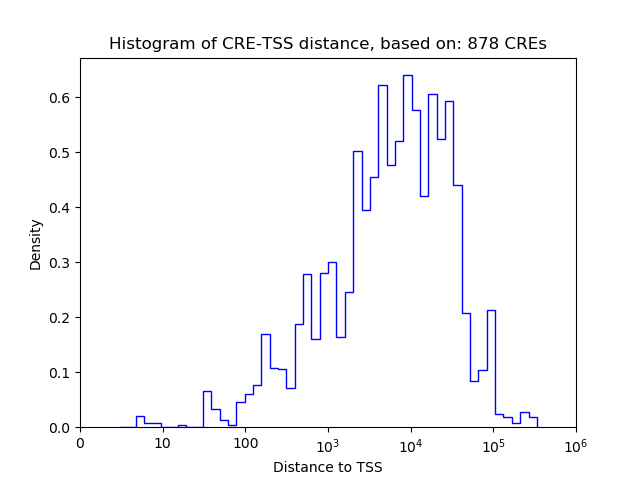
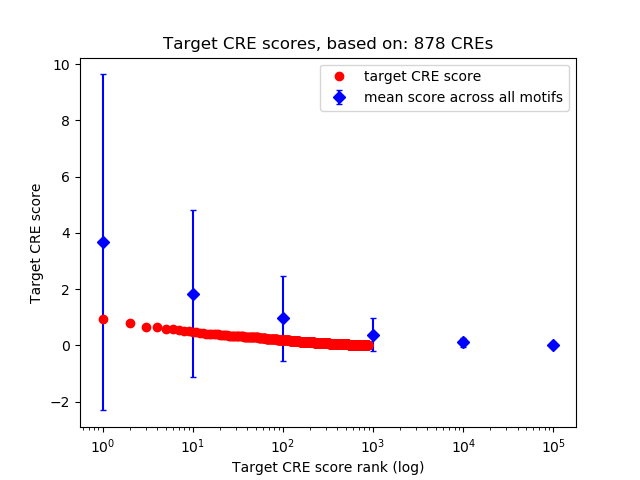
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_40160539_40160690 | 0.92 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
26672 |
0.14 |
| chr9_94704515_94704673 | 0.79 |
Gm16262 |
predicted gene 16262 |
2275 |
0.28 |
| chr13_93341639_93341827 | 0.66 |
Gm47088 |
predicted gene, 47088 |
24306 |
0.13 |
| chr8_93179883_93180034 | 0.63 |
Ces1d |
carboxylesterase 1D |
4669 |
0.15 |
| chr3_118486098_118486290 | 0.59 |
Gm26871 |
predicted gene, 26871 |
28535 |
0.12 |
| chr1_33095638_33095817 | 0.57 |
Gm7114 |
predicted gene 7114 |
5180 |
0.19 |
| chr11_16860070_16860314 | 0.54 |
Egfr |
epidermal growth factor receptor |
17958 |
0.18 |
| chr14_17696991_17697142 | 0.51 |
Thrb |
thyroid hormone receptor beta |
36170 |
0.21 |
| chr9_74892937_74893252 | 0.51 |
Onecut1 |
one cut domain, family member 1 |
26610 |
0.14 |
| chr8_35381556_35381707 | 0.48 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
4971 |
0.18 |
| chr17_62789514_62789722 | 0.46 |
Efna5 |
ephrin A5 |
91526 |
0.1 |
| chr10_87935892_87936043 | 0.44 |
Tyms-ps |
thymidylate synthase, pseudogene |
30880 |
0.15 |
| chr8_45671495_45671652 | 0.43 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
9577 |
0.22 |
| chr6_116054559_116054710 | 0.42 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
2321 |
0.27 |
| chr6_140637709_140637860 | 0.41 |
Aebp2 |
AE binding protein 2 |
32 |
0.98 |
| chr11_117278429_117279048 | 0.41 |
Septin9 |
septin 9 |
12492 |
0.18 |
| chr2_59160190_59160538 | 0.40 |
Ccdc148 |
coiled-coil domain containing 148 |
170 |
0.8 |
| chr2_178555160_178555311 | 0.40 |
Cdh26 |
cadherin-like 26 |
94605 |
0.08 |
| chr17_28007798_28007964 | 0.40 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
536 |
0.66 |
| chr9_58173012_58173189 | 0.37 |
Islr |
immunoglobulin superfamily containing leucine-rich repeat |
13879 |
0.1 |
| chr11_95385743_95385947 | 0.36 |
Slc35b1 |
solute carrier family 35, member B1 |
939 |
0.38 |
| chr4_94979002_94979670 | 0.36 |
Gm12694 |
predicted gene 12694 |
133 |
0.64 |
| chr12_32770754_32771054 | 0.35 |
Nampt |
nicotinamide phosphoribosyltransferase |
48641 |
0.13 |
| chr6_14900560_14901049 | 0.35 |
Foxp2 |
forkhead box P2 |
545 |
0.87 |
| chr13_74208331_74208542 | 0.34 |
Exoc3 |
exocyst complex component 3 |
174 |
0.95 |
| chr15_100306256_100306443 | 0.34 |
Mettl7a1 |
methyltransferase like 7A1 |
1178 |
0.33 |
| chr9_122177892_122178054 | 0.34 |
Ano10 |
anoctamin 10 |
9646 |
0.13 |
| chr8_83590177_83590330 | 0.34 |
Tecr |
trans-2,3-enoyl-CoA reductase |
4168 |
0.11 |
| chr4_136511547_136511765 | 0.33 |
Luzp1 |
leucine zipper protein 1 |
9739 |
0.16 |
| chr7_143754683_143755199 | 0.33 |
Osbpl5 |
oxysterol binding protein-like 5 |
2044 |
0.2 |
| chr1_136626185_136626488 | 0.33 |
Zfp281 |
zinc finger protein 281 |
1435 |
0.27 |
| chr3_28703876_28704177 | 0.33 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
6009 |
0.18 |
| chr9_90265733_90266048 | 0.33 |
Tbc1d2b |
TBC1 domain family, member 2B |
4879 |
0.19 |
| chr4_108362975_108363166 | 0.33 |
Shisal2a |
shisa like 2A |
20279 |
0.11 |
| chr19_24885631_24885796 | 0.32 |
Gm10053 |
predicted gene 10053 |
10027 |
0.13 |
| chr19_20612745_20612960 | 0.32 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
10891 |
0.21 |
| chr4_152229764_152230378 | 0.31 |
Gm13096 |
predicted gene 13096 |
26145 |
0.1 |
| chr5_145220307_145220458 | 0.31 |
Zfp655 |
zinc finger protein 655 |
11333 |
0.1 |
| chr6_54048004_54048155 | 0.31 |
Chn2 |
chimerin 2 |
7993 |
0.22 |
| chr15_80836341_80836511 | 0.30 |
Tnrc6b |
trinucleotide repeat containing 6b |
37711 |
0.15 |
| chr2_31493649_31493953 | 0.30 |
Ass1 |
argininosuccinate synthetase 1 |
3971 |
0.22 |
| chr13_102569889_102570107 | 0.30 |
Gm29927 |
predicted gene, 29927 |
20287 |
0.22 |
| chr5_123024278_123024718 | 0.29 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
9160 |
0.09 |
| chr3_51251222_51251373 | 0.29 |
Noct |
nocturnin |
7889 |
0.13 |
| chr19_5822213_5822895 | 0.29 |
Gm27702 |
predicted gene, 27702 |
2153 |
0.13 |
| chr4_119110584_119110758 | 0.29 |
Slc2a1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
1757 |
0.21 |
| chr13_52060964_52061138 | 0.29 |
Gm48190 |
predicted gene, 48190 |
35591 |
0.15 |
| chr7_120147685_120147843 | 0.29 |
Zp2 |
zona pellucida glycoprotein 2 |
2473 |
0.2 |
| chr11_60241446_60241847 | 0.29 |
Gm12265 |
predicted gene 12265 |
836 |
0.46 |
| chr2_66780995_66781162 | 0.29 |
Scn7a |
sodium channel, voltage-gated, type VII, alpha |
3836 |
0.32 |
| chr3_108478211_108478362 | 0.29 |
5330417C22Rik |
RIKEN cDNA 5330417C22 gene |
8205 |
0.09 |
| chr8_125474967_125475128 | 0.28 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
17663 |
0.26 |
| chr11_117072500_117072662 | 0.28 |
Snhg20 |
small nucleolar RNA host gene 20 |
3427 |
0.12 |
| chr9_9199655_9199806 | 0.27 |
Gm16833 |
predicted gene, 16833 |
36558 |
0.18 |
| chr14_75753355_75753582 | 0.27 |
Cog3 |
component of oligomeric golgi complex 3 |
1090 |
0.45 |
| chr1_74083149_74083300 | 0.27 |
Tns1 |
tensin 1 |
8127 |
0.19 |
| chr11_16851382_16851732 | 0.27 |
Egfros |
epidermal growth factor receptor, opposite strand |
20855 |
0.18 |
| chr17_84287914_84288065 | 0.26 |
Gm24492 |
predicted gene, 24492 |
242 |
0.93 |
| chr3_36533351_36533502 | 0.26 |
Gm11549 |
predicted gene 11549 |
1042 |
0.38 |
| chr1_136625692_136626183 | 0.26 |
Zfp281 |
zinc finger protein 281 |
1036 |
0.34 |
| chr4_134151936_134152400 | 0.26 |
Cep85 |
centrosomal protein 85 |
2315 |
0.16 |
| chr10_66929068_66929292 | 0.26 |
Gm26576 |
predicted gene, 26576 |
8878 |
0.16 |
| chr12_100341684_100341865 | 0.25 |
Ttc7b |
tetratricopeptide repeat domain 7B |
3478 |
0.22 |
| chr18_15332801_15332952 | 0.25 |
A830021F12Rik |
RIKEN cDNA A830021F12 gene |
19058 |
0.18 |
| chr14_121894053_121894204 | 0.24 |
1810041H14Rik |
RIKEN cDNA 1810041H14 gene |
14719 |
0.15 |
| chr2_52437849_52438032 | 0.24 |
A430018G15Rik |
RIKEN cDNA A430018G15 gene |
12928 |
0.17 |
| chr10_4781737_4781912 | 0.24 |
Esr1 |
estrogen receptor 1 (alpha) |
69173 |
0.13 |
| chr17_31180257_31180431 | 0.24 |
Tff1 |
trefoil factor 1 |
15067 |
0.1 |
| chr6_86523276_86523978 | 0.24 |
1600020E01Rik |
RIKEN cDNA 1600020E01 gene |
2620 |
0.12 |
| chr1_51891769_51891920 | 0.23 |
Myo1b |
myosin IB |
4457 |
0.17 |
| chr3_82022943_82023211 | 0.23 |
Gucy1b1 |
guanylate cyclase 1, soluble, beta 1 |
12245 |
0.17 |
| chr2_113545947_113546266 | 0.23 |
Gm13964 |
predicted gene 13964 |
42072 |
0.15 |
| chr14_32720595_32720754 | 0.23 |
Gm28651 |
predicted gene 28651 |
34116 |
0.14 |
| chr10_95255137_95255325 | 0.23 |
Gm48880 |
predicted gene, 48880 |
59622 |
0.08 |
| chr10_61443846_61444148 | 0.22 |
Gm48086 |
predicted gene, 48086 |
6720 |
0.11 |
| chr7_112271614_112271811 | 0.22 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
413 |
0.91 |
| chrX_169990489_169990640 | 0.22 |
Gm15247 |
predicted gene 15247 |
3625 |
0.18 |
| chrY_90764765_90764916 | 0.22 |
Gm21860 |
predicted gene, 21860 |
9373 |
0.17 |
| chr10_111367147_111367312 | 0.22 |
Gm40761 |
predicted gene, 40761 |
30105 |
0.16 |
| chr6_49519362_49519513 | 0.21 |
Gm5305 |
predicted gene 5305 |
24290 |
0.2 |
| chr3_51247658_51247809 | 0.21 |
Noct |
nocturnin |
4325 |
0.15 |
| chr8_111709036_111709217 | 0.21 |
Ctrb1 |
chymotrypsinogen B1 |
18116 |
0.14 |
| chr7_25888092_25888262 | 0.21 |
Gm6434 |
predicted gene 6434 |
5788 |
0.11 |
| chr5_66042796_66042956 | 0.21 |
Rbm47 |
RNA binding motif protein 47 |
11676 |
0.12 |
| chr15_59024907_59025221 | 0.21 |
Mtss1 |
MTSS I-BAR domain containing 1 |
15532 |
0.19 |
| chr19_39212535_39212719 | 0.21 |
Cyp2c53-ps |
cytochrome P450, family 2, subfamily c, polypeptide 53-ps |
16627 |
0.22 |
| chr14_71963090_71963264 | 0.20 |
4930434J06Rik |
RIKEN cDNA 4930434J06 gene |
50086 |
0.18 |
| chr3_98427563_98428016 | 0.20 |
Gm43190 |
predicted gene 43190 |
21187 |
0.13 |
| chr3_116204045_116204241 | 0.20 |
Gm31651 |
predicted gene, 31651 |
3735 |
0.2 |
| chr14_27123612_27123804 | 0.20 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
8809 |
0.22 |
| chr2_147808319_147808470 | 0.20 |
Gm25516 |
predicted gene, 25516 |
66484 |
0.11 |
| chr6_138164041_138164201 | 0.20 |
Mgst1 |
microsomal glutathione S-transferase 1 |
21267 |
0.25 |
| chrY_897914_898312 | 0.20 |
Kdm5d |
lysine (K)-specific demethylase 5D |
81 |
0.97 |
| chr7_35048630_35048913 | 0.20 |
Cebpg |
CCAAT/enhancer binding protein (C/EBP), gamma |
2734 |
0.12 |
| chr11_84290556_84290741 | 0.20 |
Acaca |
acetyl-Coenzyme A carboxylase alpha |
10193 |
0.25 |
| chr1_165199539_165199690 | 0.20 |
Sft2d2 |
SFT2 domain containing 2 |
5176 |
0.16 |
| chr17_87571110_87571333 | 0.20 |
Gm46587 |
predicted gene, 46587 |
433 |
0.82 |
| chr10_69247438_69247589 | 0.20 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
18124 |
0.18 |
| chr7_104231335_104231499 | 0.20 |
Trim6 |
tripartite motif-containing 6 |
5910 |
0.08 |
| chr3_31107496_31107647 | 0.19 |
Skil |
SKI-like |
10738 |
0.19 |
| chr10_20174033_20174184 | 0.19 |
Map7 |
microtubule-associated protein 7 |
2101 |
0.28 |
| chr9_66199300_66199667 | 0.19 |
Dapk2 |
death-associated protein kinase 2 |
21264 |
0.19 |
| chr9_43045468_43045658 | 0.19 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
2931 |
0.29 |
| chr5_92718022_92718394 | 0.19 |
Gm20500 |
predicted gene 20500 |
17997 |
0.17 |
| chr4_97789633_97790009 | 0.19 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
11743 |
0.2 |
| chr8_47532777_47532928 | 0.19 |
Trappc11 |
trafficking protein particle complex 11 |
615 |
0.49 |
| chr6_28605878_28606042 | 0.18 |
Gm37978 |
predicted gene, 37978 |
37888 |
0.14 |
| chr13_118386920_118387073 | 0.18 |
Mrps30 |
mitochondrial ribosomal protein S30 |
256 |
0.62 |
| chr10_75514104_75514407 | 0.18 |
Gucd1 |
guanylyl cyclase domain containing 1 |
2915 |
0.15 |
| chr3_28467874_28468055 | 0.18 |
Mir466q |
microRNA 466q |
48021 |
0.15 |
| chr18_12637348_12637569 | 0.18 |
Ttc39c |
tetratricopeptide repeat domain 39C |
5886 |
0.16 |
| chr3_93520801_93521129 | 0.18 |
S100a11 |
S100 calcium binding protein A11 |
477 |
0.69 |
| chr6_108442018_108442194 | 0.17 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
16597 |
0.18 |
| chr10_20472491_20472664 | 0.17 |
Pde7b |
phosphodiesterase 7B |
25686 |
0.18 |
| chr14_62884538_62884723 | 0.17 |
n-R5s47 |
nuclear encoded rRNA 5S 47 |
14916 |
0.13 |
| chr9_35112195_35112560 | 0.17 |
4930581F22Rik |
RIKEN cDNA 4930581F22 gene |
4351 |
0.16 |
| chr8_54518624_54518791 | 0.17 |
Gm45553 |
predicted gene 45553 |
4557 |
0.2 |
| chr8_122259476_122259634 | 0.17 |
Zfp469 |
zinc finger protein 469 |
935 |
0.55 |
| chr12_8110158_8110350 | 0.17 |
Ldah |
lipid droplet associated hydrolase |
97853 |
0.06 |
| chr18_20660293_20660465 | 0.17 |
Gm16090 |
predicted gene 16090 |
4881 |
0.18 |
| chr18_38775220_38775405 | 0.17 |
Gm8302 |
predicted gene 8302 |
2965 |
0.27 |
| chr16_6366127_6366312 | 0.17 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
17081 |
0.31 |
| chr14_22633682_22633865 | 0.17 |
Lrmda |
leucine rich melanocyte differentiation associated |
37260 |
0.19 |
| chr3_22008291_22008442 | 0.17 |
Gm43674 |
predicted gene 43674 |
9898 |
0.19 |
| chr14_30930496_30930655 | 0.17 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
6815 |
0.11 |
| chr8_77199905_77200064 | 0.16 |
Gm23260 |
predicted gene, 23260 |
5289 |
0.24 |
| chr13_69488805_69488956 | 0.16 |
Gm48676 |
predicted gene, 48676 |
25120 |
0.13 |
| chr6_113078560_113078717 | 0.16 |
Setd5 |
SET domain containing 5 |
999 |
0.33 |
| chr2_178684195_178684361 | 0.16 |
Cdh26 |
cadherin-like 26 |
223648 |
0.02 |
| chr7_51972606_51972763 | 0.16 |
Gas2 |
growth arrest specific 2 |
29000 |
0.14 |
| chr10_15746708_15746902 | 0.16 |
Gm32283 |
predicted gene, 32283 |
7983 |
0.18 |
| chr6_139596135_139596315 | 0.16 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
3255 |
0.26 |
| chr8_105050817_105050971 | 0.16 |
Gm45724 |
predicted gene 45724 |
574 |
0.57 |
| chr8_80493229_80493398 | 0.16 |
Gypa |
glycophorin A |
468 |
0.85 |
| chr4_122981345_122981517 | 0.15 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
14221 |
0.13 |
| chr17_47009670_47009958 | 0.15 |
Ubr2 |
ubiquitin protein ligase E3 component n-recognin 2 |
665 |
0.67 |
| chr14_48332738_48332892 | 0.15 |
1700128I11Rik |
RIKEN cDNA 1700128I11 gene |
29800 |
0.13 |
| chr2_167830636_167830820 | 0.15 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
2918 |
0.22 |
| chr11_118201294_118201477 | 0.15 |
Gm11737 |
predicted gene 11737 |
3794 |
0.18 |
| chr17_3244258_3244409 | 0.15 |
Gm49797 |
predicted gene, 49797 |
13062 |
0.17 |
| chr10_68092838_68093042 | 0.14 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
43686 |
0.14 |
| chr19_55113023_55113249 | 0.14 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
10045 |
0.2 |
| chr9_62143745_62144052 | 0.14 |
Glce |
glucuronyl C5-epimerase |
21243 |
0.17 |
| chr12_16585048_16585237 | 0.14 |
Lpin1 |
lipin 1 |
4578 |
0.27 |
| chr9_48717198_48717480 | 0.14 |
Nnmt |
nicotinamide N-methyltransferase |
112186 |
0.06 |
| chr11_76178910_76179262 | 0.14 |
Vps53 |
VPS53 GARP complex subunit |
413 |
0.56 |
| chr2_103424741_103424905 | 0.14 |
Elf5 |
E74-like factor 5 |
5 |
0.98 |
| chr7_100993375_100993782 | 0.14 |
P2ry2 |
purinergic receptor P2Y, G-protein coupled 2 |
10309 |
0.14 |
| chr3_51227403_51227633 | 0.14 |
Noct |
nocturnin |
3048 |
0.19 |
| chr13_49653443_49653648 | 0.14 |
Nol8 |
nucleolar protein 8 |
195 |
0.83 |
| chr11_97850871_97851022 | 0.14 |
Fbxo47 |
F-box protein 47 |
4606 |
0.11 |
| chr2_58784544_58784695 | 0.14 |
Upp2 |
uridine phosphorylase 2 |
19294 |
0.19 |
| chr6_122596666_122596866 | 0.13 |
Apobec1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
3361 |
0.14 |
| chr5_40741917_40742189 | 0.13 |
Gm23022 |
predicted gene, 23022 |
336124 |
0.01 |
| chr2_62362282_62362458 | 0.13 |
Dpp4 |
dipeptidylpeptidase 4 |
4663 |
0.2 |
| chr2_148686320_148686544 | 0.13 |
Gzf1 |
GDNF-inducible zinc finger protein 1 |
1820 |
0.28 |
| chr15_58592792_58592961 | 0.13 |
Fer1l6 |
fer-1-like 6 (C. elegans) |
45629 |
0.16 |
| chr16_42907740_42907907 | 0.13 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
171 |
0.95 |
| chr8_91378014_91378400 | 0.13 |
Fto |
fat mass and obesity associated |
12348 |
0.15 |
| chr16_93342574_93342799 | 0.13 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
10503 |
0.18 |
| chr3_145655180_145655331 | 0.13 |
Gm17501 |
predicted gene, 17501 |
3161 |
0.24 |
| chr2_104815506_104815833 | 0.13 |
Qser1 |
glutamine and serine rich 1 |
1027 |
0.45 |
| chr4_62010687_62011047 | 0.13 |
Mup-ps20 |
major urinary protein, pseudogene 20 |
3990 |
0.18 |
| chr2_27575873_27576343 | 0.13 |
Gm13421 |
predicted gene 13421 |
35679 |
0.13 |
| chr1_36100470_36100674 | 0.13 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
1118 |
0.4 |
| chr1_153702024_153702276 | 0.13 |
Gm29529 |
predicted gene 29529 |
9494 |
0.11 |
| chr16_46002412_46002585 | 0.12 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
7720 |
0.17 |
| chr6_140423640_140424226 | 0.12 |
Plekha5 |
pleckstrin homology domain containing, family A member 5 |
121 |
0.97 |
| chr9_120582294_120582487 | 0.12 |
Gm47062 |
predicted gene, 47062 |
2255 |
0.19 |
| chr4_40221217_40221381 | 0.12 |
Ddx58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
699 |
0.66 |
| chr7_137288505_137288677 | 0.12 |
Ebf3 |
early B cell factor 3 |
21053 |
0.18 |
| chr18_74646609_74646944 | 0.12 |
Myo5b |
myosin VB |
13599 |
0.23 |
| chr18_64377874_64378033 | 0.12 |
Onecut2 |
one cut domain, family member 2 |
37933 |
0.12 |
| chr4_62051294_62051807 | 0.12 |
Mup20 |
major urinary protein 20 |
2608 |
0.2 |
| chr13_32801542_32801715 | 0.12 |
Wrnip1 |
Werner helicase interacting protein 1 |
410 |
0.72 |
| chr19_34745934_34746276 | 0.12 |
Slc16a12 |
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
1184 |
0.36 |
| chr14_76149595_76149834 | 0.12 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
38823 |
0.15 |
| chr1_67130174_67130732 | 0.12 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
7427 |
0.25 |
| chr6_59453057_59453208 | 0.12 |
Gprin3 |
GPRIN family member 3 |
26838 |
0.26 |
| chr4_45735841_45735999 | 0.12 |
Gm12410 |
predicted gene 12410 |
20751 |
0.13 |
| chr6_17282218_17282369 | 0.12 |
Cav2 |
caveolin 2 |
934 |
0.59 |
| chr15_35301858_35302156 | 0.12 |
Osr2 |
odd-skipped related 2 |
1707 |
0.37 |
| chr13_56440067_56440428 | 0.12 |
Slc25a48 |
solute carrier family 25, member 48 |
1892 |
0.29 |
| chr9_74994481_74994908 | 0.12 |
Fam214a |
family with sequence similarity 214, member A |
18583 |
0.17 |
| chr10_26855717_26855868 | 0.12 |
Arhgap18 |
Rho GTPase activating protein 18 |
7550 |
0.25 |
| chr10_20153471_20153802 | 0.12 |
Map7 |
microtubule-associated protein 7 |
4638 |
0.2 |
| chr14_19663725_19663937 | 0.12 |
Gm17030 |
predicted gene 17030 |
13829 |
0.1 |
| chr18_46595577_46595776 | 0.12 |
Tmed7 |
transmembrane p24 trafficking protein 7 |
1638 |
0.26 |
| chr6_89169156_89169504 | 0.12 |
Gm6507 |
predicted gene 6507 |
16695 |
0.14 |
| chr1_172307825_172307976 | 0.11 |
Igsf8 |
immunoglobulin superfamily, member 8 |
3871 |
0.12 |
| chr15_103215066_103215308 | 0.11 |
Cbx5 |
chromobox 5 |
118 |
0.94 |
| chr15_31237015_31237437 | 0.11 |
Dap |
death-associated protein |
12030 |
0.17 |
| chr8_117719897_117720243 | 0.11 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
866 |
0.51 |
| chr6_149096234_149096512 | 0.11 |
Dennd5b |
DENN/MADD domain containing 5B |
5165 |
0.12 |
| chr12_81190858_81191019 | 0.11 |
Mir3067 |
microRNA 3067 |
24787 |
0.17 |
| chr9_21081433_21081847 | 0.11 |
Gm38431 |
predicted gene, 38431 |
288 |
0.75 |
| chr12_3647979_3648147 | 0.11 |
Dtnb |
dystrobrevin, beta |
15141 |
0.21 |
| chr3_51231545_51231714 | 0.11 |
Gm38357 |
predicted gene, 38357 |
288 |
0.87 |
| chr12_103947018_103947616 | 0.11 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
9581 |
0.11 |
| chr2_128936241_128936451 | 0.11 |
Zc3h8 |
zinc finger CCCH type containing 8 |
7731 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |