Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
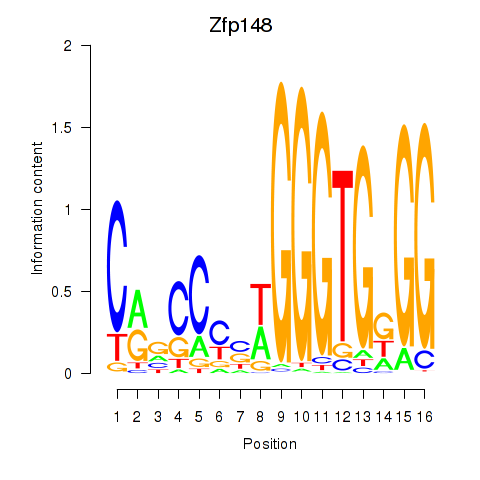
Results for Zfp148
Z-value: 1.00
Transcription factors associated with Zfp148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp148
|
ENSMUSG00000022811.10 | zinc finger protein 148 |
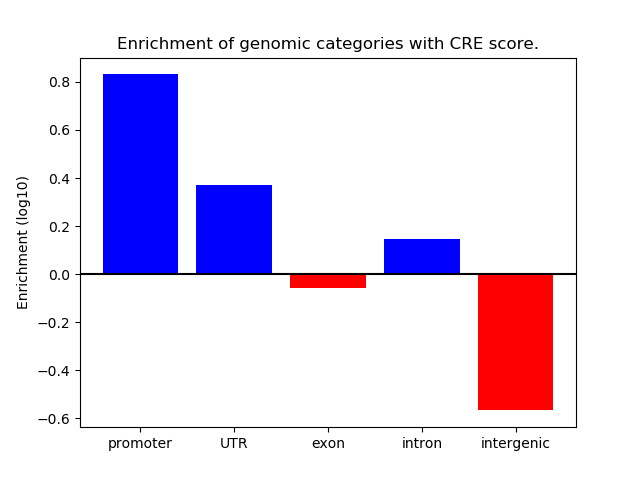
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_33394234_33394423 | Zfp148 | 6117 | 0.227114 | -0.60 | 2.1e-01 | Click! |
| chr16_33404197_33404348 | Zfp148 | 3827 | 0.264431 | 0.57 | 2.4e-01 | Click! |
| chr16_33400060_33400223 | Zfp148 | 304 | 0.915765 | -0.54 | 2.6e-01 | Click! |
| chr16_33408360_33408511 | Zfp148 | 7990 | 0.227147 | -0.45 | 3.7e-01 | Click! |
| chr16_33401041_33401192 | Zfp148 | 671 | 0.741024 | -0.43 | 4.0e-01 | Click! |
Activity of the Zfp148 motif across conditions
Conditions sorted by the z-value of the Zfp148 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
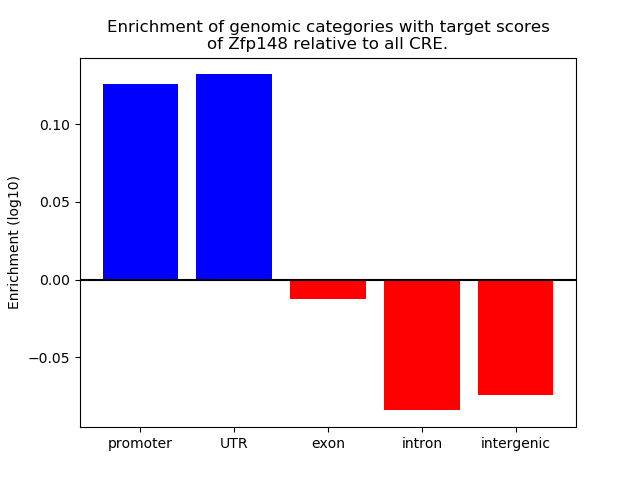
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_127341729_127341929 | 0.85 |
Gm42458 |
predicted gene 42458 |
15972 |
0.12 |
| chr9_74892937_74893252 | 0.84 |
Onecut1 |
one cut domain, family member 1 |
26610 |
0.14 |
| chr7_139388680_139389161 | 0.72 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
189 |
0.96 |
| chr1_178600841_178601326 | 0.56 |
Gm24405 |
predicted gene, 24405 |
35822 |
0.18 |
| chr9_122634913_122635137 | 0.50 |
Gm47134 |
predicted gene, 47134 |
12805 |
0.13 |
| chr7_15939282_15939433 | 0.48 |
Snord23 |
small nucleolar RNA, C/D box 23 |
487 |
0.65 |
| chr18_12947344_12947514 | 0.45 |
Osbpl1a |
oxysterol binding protein-like 1A |
5588 |
0.19 |
| chr18_76542799_76542950 | 0.44 |
Gm31933 |
predicted gene, 31933 |
89996 |
0.09 |
| chr10_86689713_86689885 | 0.43 |
Gm15344 |
predicted gene 15344 |
3296 |
0.09 |
| chr17_36867279_36867493 | 0.42 |
Trim15 |
tripartite motif-containing 15 |
176 |
0.87 |
| chr5_125524109_125525122 | 0.38 |
Tmem132b |
transmembrane protein 132B |
7159 |
0.16 |
| chr9_22070765_22071174 | 0.38 |
Gm16845 |
predicted gene, 16845 |
33 |
0.67 |
| chr15_27477536_27477721 | 0.38 |
Tiaf2 |
TGF-beta1-induced anti-apoptotic factor 2 |
1378 |
0.37 |
| chr14_118783672_118783823 | 0.37 |
Cldn10 |
claudin 10 |
4161 |
0.19 |
| chr6_35259636_35259787 | 0.37 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
6979 |
0.16 |
| chr19_22930600_22930751 | 0.36 |
Gm50136 |
predicted gene, 50136 |
130779 |
0.05 |
| chr10_80577672_80578424 | 0.34 |
Klf16 |
Kruppel-like factor 16 |
727 |
0.41 |
| chr5_144255609_144256174 | 0.33 |
2900089D17Rik |
RIKEN cDNA 2900089D17 gene |
295 |
0.71 |
| chr5_134887912_134888078 | 0.32 |
Tmem270 |
transmembrane protein 270 |
18738 |
0.09 |
| chr11_98775529_98775744 | 0.32 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
303 |
0.81 |
| chr1_20619402_20619555 | 0.31 |
Pkhd1 |
polycystic kidney and hepatic disease 1 |
1414 |
0.43 |
| chr10_127351803_127352162 | 0.31 |
Inhbe |
inhibin beta-E |
2429 |
0.13 |
| chr7_25686720_25687008 | 0.29 |
Tgfb1 |
transforming growth factor, beta 1 |
138 |
0.91 |
| chr10_128746357_128746529 | 0.29 |
Pym1 |
PYM homolog 1, exon junction complex associated factor |
1446 |
0.19 |
| chr11_3332327_3332681 | 0.29 |
Pik3ip1 |
phosphoinositide-3-kinase interacting protein 1 |
53 |
0.94 |
| chr4_117997004_117997205 | 0.29 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
22068 |
0.14 |
| chr5_52613790_52613948 | 0.29 |
8030423F21Rik |
RIKEN cDNA 8030423F21 gene |
5143 |
0.18 |
| chr1_21264707_21265188 | 0.29 |
Gm28836 |
predicted gene 28836 |
6646 |
0.11 |
| chr5_125528257_125528432 | 0.29 |
Tmem132b |
transmembrane protein 132B |
3430 |
0.21 |
| chr3_142745096_142745348 | 0.29 |
Gm15540 |
predicted gene 15540 |
541 |
0.67 |
| chr19_37434647_37435026 | 0.28 |
Hhex |
hematopoietically expressed homeobox |
26 |
0.69 |
| chr2_25145394_25145761 | 0.28 |
Gm13387 |
predicted gene 13387 |
181 |
0.87 |
| chr19_10101442_10101987 | 0.28 |
Fads2 |
fatty acid desaturase 2 |
32 |
0.97 |
| chr7_144942500_144942684 | 0.28 |
Ccnd1 |
cyclin D1 |
2667 |
0.2 |
| chr9_105465998_105466149 | 0.28 |
Atp2c1 |
ATPase, Ca++-sequestering |
4317 |
0.19 |
| chr1_168281652_168281964 | 0.28 |
Gm37524 |
predicted gene, 37524 |
55863 |
0.14 |
| chr4_139631622_139632150 | 0.28 |
Aldh4a1 |
aldehyde dehydrogenase 4 family, member A1 |
8777 |
0.14 |
| chr14_117326010_117326161 | 0.27 |
Gpc6 |
glypican 6 |
400149 |
0.01 |
| chr6_82760855_82761006 | 0.27 |
Gm17034 |
predicted gene 17034 |
3352 |
0.19 |
| chr1_172274690_172274870 | 0.27 |
Atp1a2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
1698 |
0.22 |
| chr6_15777156_15777307 | 0.27 |
Gm43924 |
predicted gene, 43924 |
34092 |
0.18 |
| chr5_147321463_147321654 | 0.27 |
Urad |
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
882 |
0.42 |
| chr11_98839480_98839639 | 0.26 |
Rapgefl1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
2774 |
0.15 |
| chr19_14586215_14586387 | 0.26 |
Tle4 |
transducin-like enhancer of split 4 |
9238 |
0.28 |
| chr5_135719979_135720130 | 0.26 |
Por |
P450 (cytochrome) oxidoreductase |
4049 |
0.12 |
| chr3_79994107_79994258 | 0.25 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
60710 |
0.11 |
| chr5_122109359_122109528 | 0.25 |
Ccdc63 |
coiled-coil domain containing 63 |
1670 |
0.28 |
| chr6_91736603_91736766 | 0.25 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
2965 |
0.19 |
| chr14_47534004_47534155 | 0.25 |
Gm49004 |
predicted gene, 49004 |
7901 |
0.12 |
| chr4_138979154_138979535 | 0.25 |
Tmco4 |
transmembrane and coiled-coil domains 4 |
6435 |
0.16 |
| chr9_107307749_107307922 | 0.25 |
Gm17041 |
predicted gene 17041 |
5997 |
0.1 |
| chr4_139629117_139629268 | 0.25 |
Aldh4a1 |
aldehyde dehydrogenase 4 family, member A1 |
6083 |
0.15 |
| chr14_26454997_26455164 | 0.25 |
Slmap |
sarcolemma associated protein |
5092 |
0.18 |
| chr16_17924359_17924530 | 0.24 |
AA914427 |
EST AA914427 |
2401 |
0.13 |
| chr4_135989135_135989286 | 0.24 |
Pithd1 |
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
1946 |
0.16 |
| chr16_34925073_34925241 | 0.24 |
Mylk |
myosin, light polypeptide kinase |
5185 |
0.21 |
| chr5_57743122_57743273 | 0.24 |
Gm42635 |
predicted gene 42635 |
18804 |
0.12 |
| chr5_33053298_33053472 | 0.24 |
Gm43849 |
predicted gene 43849 |
3400 |
0.18 |
| chr12_16753229_16753413 | 0.24 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
3874 |
0.2 |
| chr1_170997840_170997991 | 0.24 |
Gm10522 |
predicted gene 10522 |
15971 |
0.08 |
| chr2_30547946_30548102 | 0.23 |
Gm14487 |
predicted gene 14487 |
13295 |
0.16 |
| chr13_107469451_107469885 | 0.23 |
AI197445 |
expressed sequence AI197445 |
154 |
0.97 |
| chr10_127577568_127577719 | 0.23 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
4242 |
0.13 |
| chr8_117548542_117548713 | 0.23 |
Gm27361 |
predicted gene, 27361 |
1601 |
0.41 |
| chr18_38406873_38407073 | 0.23 |
Ndfip1 |
Nedd4 family interacting protein 1 |
3423 |
0.18 |
| chr4_155894332_155894496 | 0.23 |
Acap3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
2102 |
0.12 |
| chr2_27595201_27595352 | 0.23 |
Gm13421 |
predicted gene 13421 |
54847 |
0.1 |
| chr12_104882664_104882819 | 0.22 |
4930408O17Rik |
RIKEN cDNA 4930408O17 gene |
17411 |
0.16 |
| chr15_98734151_98734335 | 0.22 |
Fkbp11 |
FK506 binding protein 11 |
6045 |
0.1 |
| chr9_119475158_119475309 | 0.22 |
Exog |
endo/exonuclease (5'-3'), endonuclease G-like |
30246 |
0.12 |
| chr14_75055064_75055264 | 0.22 |
Rubcnl |
RUN and cysteine rich domain containing beclin 1 interacting protein like |
18542 |
0.18 |
| chr15_25622272_25622843 | 0.22 |
Myo10 |
myosin X |
8 |
0.98 |
| chr8_81341615_81341766 | 0.22 |
Inpp4b |
inositol polyphosphate-4-phosphatase, type II |
866 |
0.66 |
| chr10_86793800_86793963 | 0.22 |
Nt5dc3 |
5'-nucleotidase domain containing 3 |
14876 |
0.11 |
| chr6_88299541_88299692 | 0.22 |
Gm44265 |
predicted gene, 44265 |
53990 |
0.09 |
| chr15_100100443_100100600 | 0.21 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
7640 |
0.15 |
| chr6_91525048_91525199 | 0.21 |
Gm45216 |
predicted gene 45216 |
2714 |
0.17 |
| chr5_35033214_35033391 | 0.21 |
Gm43791 |
predicted gene 43791 |
143 |
0.94 |
| chr11_65002970_65003224 | 0.20 |
Elac2 |
elaC ribonuclease Z 2 |
2761 |
0.28 |
| chr5_112242342_112242634 | 0.20 |
Cryba4 |
crystallin, beta A4 |
8674 |
0.11 |
| chr9_107315743_107315923 | 0.20 |
Hemk1 |
HemK methyltransferase family member 1 |
13619 |
0.09 |
| chr4_141873016_141873167 | 0.20 |
Efhd2 |
EF hand domain containing 2 |
1829 |
0.2 |
| chr19_10634110_10634499 | 0.20 |
Vwce |
von Willebrand factor C and EGF domains |
22 |
0.95 |
| chr18_75380339_75380510 | 0.20 |
Smad7 |
SMAD family member 7 |
5510 |
0.23 |
| chr11_90389588_90389755 | 0.20 |
Hlf |
hepatic leukemia factor |
19 |
0.99 |
| chr12_76927037_76927196 | 0.20 |
Max |
Max protein |
12878 |
0.15 |
| chr1_151500754_151501127 | 0.20 |
Rnf2 |
ring finger protein 2 |
15 |
0.95 |
| chr19_4462072_4462253 | 0.20 |
Syt12 |
synaptotagmin XII |
10011 |
0.12 |
| chr8_105090572_105090895 | 0.20 |
Ces3b |
carboxylesterase 3B |
2114 |
0.18 |
| chr18_4691956_4692115 | 0.19 |
Jcad |
junctional cadherin 5 associated |
42967 |
0.15 |
| chr16_35635109_35635270 | 0.19 |
Sema5b |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
22906 |
0.17 |
| chr7_4504776_4504978 | 0.19 |
Tnnt1 |
troponin T1, skeletal, slow |
3148 |
0.1 |
| chr9_62491243_62491394 | 0.19 |
Coro2b |
coronin, actin binding protein, 2B |
19180 |
0.2 |
| chr6_128090958_128091168 | 0.19 |
Gm26338 |
predicted gene, 26338 |
3963 |
0.19 |
| chr5_111378490_111378641 | 0.19 |
Pitpnb |
phosphatidylinositol transfer protein, beta |
1604 |
0.34 |
| chr14_74924136_74924288 | 0.19 |
Gm10847 |
predicted gene 10847 |
2233 |
0.32 |
| chr16_20716096_20716505 | 0.19 |
Clcn2 |
chloride channel, voltage-sensitive 2 |
136 |
0.88 |
| chr8_95884773_95884931 | 0.19 |
Got2 |
glutamatic-oxaloacetic transaminase 2, mitochondrial |
3540 |
0.14 |
| chr6_88292396_88292777 | 0.19 |
Gm44265 |
predicted gene, 44265 |
61020 |
0.08 |
| chr17_47304315_47304500 | 0.19 |
Trerf1 |
transcriptional regulating factor 1 |
10338 |
0.16 |
| chr10_23894548_23894745 | 0.19 |
Vnn1 |
vanin 1 |
42 |
0.96 |
| chr11_98768600_98768751 | 0.18 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
1751 |
0.2 |
| chr10_69910495_69910908 | 0.18 |
Ank3 |
ankyrin 3, epithelial |
4164 |
0.35 |
| chr11_98785850_98786019 | 0.18 |
Msl1 |
male specific lethal 1 |
9582 |
0.09 |
| chr4_129810784_129811411 | 0.18 |
Ptp4a2 |
protein tyrosine phosphatase 4a2 |
122 |
0.95 |
| chr3_65658910_65659081 | 0.18 |
Mir8120 |
microRNA 8120 |
293 |
0.87 |
| chr18_12663657_12663965 | 0.18 |
Gm41668 |
predicted gene, 41668 |
15392 |
0.14 |
| chr7_66689696_66689866 | 0.18 |
Lins1 |
lines homolog 1 |
108 |
0.61 |
| chr12_112593582_112593778 | 0.18 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
4414 |
0.16 |
| chr9_108820189_108820340 | 0.18 |
Gm35025 |
predicted gene, 35025 |
4555 |
0.09 |
| chr9_43744780_43745133 | 0.18 |
Nectin1 |
nectin cell adhesion molecule 1 |
312 |
0.63 |
| chrX_140541717_140541904 | 0.18 |
Tsc22d3 |
TSC22 domain family, member 3 |
858 |
0.63 |
| chr13_48260781_48261195 | 0.18 |
Id4 |
inhibitor of DNA binding 4 |
240 |
0.85 |
| chr2_32448342_32448493 | 0.17 |
Naif1 |
nuclear apoptosis inducing factor 1 |
2040 |
0.18 |
| chr1_45904282_45904665 | 0.17 |
Gm18303 |
predicted gene, 18303 |
1150 |
0.39 |
| chr7_141193568_141193999 | 0.17 |
Hras |
Harvey rat sarcoma virus oncogene |
99 |
0.77 |
| chr19_26746874_26747161 | 0.17 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
583 |
0.8 |
| chr5_34346535_34346723 | 0.17 |
Rnf4 |
ring finger protein 4 |
4487 |
0.15 |
| chr9_122635651_122636097 | 0.17 |
Gm47134 |
predicted gene, 47134 |
11956 |
0.13 |
| chr11_83596101_83596252 | 0.17 |
Ccl6 |
chemokine (C-C motif) ligand 6 |
3089 |
0.14 |
| chr4_136090324_136090475 | 0.17 |
Gm13009 |
predicted gene 13009 |
15427 |
0.12 |
| chr1_55052302_55052488 | 0.17 |
Coq10b |
coenzyme Q10B |
375 |
0.63 |
| chr5_74738458_74738660 | 0.17 |
Gm15985 |
predicted gene 15985 |
4108 |
0.2 |
| chr11_55461149_55461526 | 0.17 |
Atox1 |
antioxidant 1 copper chaperone |
98 |
0.94 |
| chr9_107660530_107661377 | 0.17 |
Slc38a3 |
solute carrier family 38, member 3 |
1425 |
0.2 |
| chr2_4508012_4508163 | 0.17 |
Frmd4a |
FERM domain containing 4A |
34893 |
0.15 |
| chr14_59219365_59219526 | 0.17 |
Rcbtb1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
1840 |
0.33 |
| chr9_65679332_65679533 | 0.17 |
Oaz2 |
ornithine decarboxylase antizyme 2 |
2841 |
0.22 |
| chr7_45705992_45706174 | 0.17 |
Dbp |
D site albumin promoter binding protein |
799 |
0.31 |
| chr5_146247315_146247530 | 0.17 |
Gm15739 |
predicted gene 15739 |
1129 |
0.38 |
| chr2_152925622_152925788 | 0.17 |
Foxs1 |
forkhead box S1 |
7503 |
0.12 |
| chr3_90615494_90615652 | 0.17 |
S100a6 |
S100 calcium binding protein A6 (calcyclin) |
1813 |
0.16 |
| chr7_132713503_132713681 | 0.17 |
Gm15582 |
predicted gene 15582 |
53708 |
0.1 |
| chr11_69996851_69997024 | 0.17 |
Phf23 |
PHD finger protein 23 |
4 |
0.92 |
| chr6_37870282_37871066 | 0.16 |
Trim24 |
tripartite motif-containing 24 |
137 |
0.96 |
| chr11_98770698_98771034 | 0.16 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
440 |
0.69 |
| chr5_73993652_73993812 | 0.16 |
Usp46os1 |
ubiquitin specific peptidase 46, opposite strand 1 |
20483 |
0.13 |
| chr7_26304515_26304666 | 0.16 |
Cyp2a4 |
cytochrome P450, family 2, subfamily a, polypeptide 4 |
2579 |
0.18 |
| chr14_45452270_45452458 | 0.16 |
Gm34250 |
predicted gene, 34250 |
5291 |
0.14 |
| chr8_25806161_25806351 | 0.16 |
Star |
steroidogenic acute regulatory protein |
299 |
0.83 |
| chr13_31558063_31558442 | 0.16 |
A530084C06Rik |
RIKEN cDNA A530084C06 gene |
1081 |
0.38 |
| chr11_106263631_106263795 | 0.16 |
Smarcd2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
1223 |
0.25 |
| chr5_135161333_135161493 | 0.16 |
Bcl7b |
B cell CLL/lymphoma 7B |
6870 |
0.11 |
| chr15_76195723_76195905 | 0.16 |
Plec |
plectin |
104 |
0.92 |
| chr16_38376538_38376695 | 0.16 |
Popdc2 |
popeye domain containing 2 |
4618 |
0.14 |
| chr15_59062348_59062499 | 0.16 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6959 |
0.24 |
| chr10_89460241_89460393 | 0.16 |
Gas2l3 |
growth arrest-specific 2 like 3 |
16350 |
0.22 |
| chr11_100398084_100398235 | 0.16 |
Jup |
junction plakoglobin |
396 |
0.67 |
| chr11_16813698_16813849 | 0.16 |
Egfros |
epidermal growth factor receptor, opposite strand |
16929 |
0.21 |
| chr14_62761438_62761880 | 0.16 |
Ints6 |
integrator complex subunit 6 |
490 |
0.76 |
| chr2_30185581_30185818 | 0.15 |
Spout1 |
SPOUT domain containing methyltransferase 1 |
7240 |
0.1 |
| chr17_78417642_78418010 | 0.15 |
Fez2 |
fasciculation and elongation protein zeta 2 (zygin II) |
135 |
0.95 |
| chr13_9145204_9145368 | 0.15 |
Gm28155 |
predicted gene 28155 |
4405 |
0.19 |
| chr11_16862611_16863177 | 0.15 |
Egfr |
epidermal growth factor receptor |
15256 |
0.19 |
| chr10_95508892_95509043 | 0.15 |
Ube2n |
ubiquitin-conjugating enzyme E2N |
6178 |
0.13 |
| chr12_86836691_86837004 | 0.15 |
Gm10095 |
predicted gene 10095 |
9620 |
0.19 |
| chr2_181918621_181918792 | 0.15 |
Gm5466 |
predicted gene 5466 |
21883 |
0.17 |
| chr7_19928412_19928563 | 0.15 |
Pvr |
poliovirus receptor |
7327 |
0.08 |
| chr1_133363345_133363819 | 0.15 |
Etnk2 |
ethanolamine kinase 2 |
6 |
0.97 |
| chr8_95888030_95888197 | 0.14 |
Got2 |
glutamatic-oxaloacetic transaminase 2, mitochondrial |
279 |
0.85 |
| chr9_75040383_75040575 | 0.14 |
Arpp19 |
cAMP-regulated phosphoprotein 19 |
2562 |
0.24 |
| chr15_58995280_58995901 | 0.14 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
11454 |
0.17 |
| chr7_89558667_89558818 | 0.14 |
Gm45064 |
predicted gene 45064 |
6376 |
0.17 |
| chr14_25207344_25207704 | 0.14 |
4930572O13Rik |
RIKEN cDNA 4930572O13 gene |
64283 |
0.1 |
| chr7_133315419_133315795 | 0.14 |
Gm45672 |
predicted gene 45672 |
17594 |
0.25 |
| chr4_3373410_3373589 | 0.14 |
Gm11784 |
predicted gene 11784 |
13347 |
0.2 |
| chr13_47043121_47043523 | 0.14 |
Tpmt |
thiopurine methyltransferase |
21 |
0.68 |
| chr7_140851973_140852179 | 0.14 |
Bet1l |
Bet1 golgi vesicular membrane trafficking protein like |
4170 |
0.07 |
| chr17_28495920_28496094 | 0.14 |
Fkbp5 |
FK506 binding protein 5 |
9858 |
0.08 |
| chr15_27496291_27496442 | 0.14 |
B230362B09Rik |
RIKEN cDNA B230362B09 gene |
7546 |
0.17 |
| chr3_137940872_137941024 | 0.14 |
Dapp1 |
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
13765 |
0.1 |
| chr3_89378414_89378785 | 0.14 |
Zbtb7b |
zinc finger and BTB domain containing 7B |
8590 |
0.06 |
| chr7_118250628_118250799 | 0.14 |
4930583K01Rik |
RIKEN cDNA 4930583K01 gene |
6848 |
0.14 |
| chr14_7974818_7975082 | 0.14 |
Dnase1l3 |
deoxyribonuclease 1-like 3 |
2272 |
0.27 |
| chr8_90869900_90870054 | 0.14 |
Gm45640 |
predicted gene 45640 |
6604 |
0.14 |
| chr1_177495025_177495183 | 0.14 |
Gm37306 |
predicted gene, 37306 |
27726 |
0.14 |
| chr19_6415578_6415765 | 0.14 |
Gm14965 |
predicted gene 14965 |
2935 |
0.12 |
| chr18_84087281_84087954 | 0.13 |
Tshz1 |
teashirt zinc finger family member 1 |
89 |
0.85 |
| chr2_59670423_59670592 | 0.13 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
23696 |
0.24 |
| chr8_121980914_121981086 | 0.13 |
Banp |
BTG3 associated nuclear protein |
2678 |
0.16 |
| chr4_150909812_150910129 | 0.13 |
Park7 |
Parkinson disease (autosomal recessive, early onset) 7 |
44 |
0.97 |
| chr1_36095531_36095713 | 0.13 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
6068 |
0.15 |
| chr5_111509539_111510088 | 0.13 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
71609 |
0.09 |
| chr1_82554461_82554615 | 0.13 |
Col4a4 |
collagen, type IV, alpha 4 |
32254 |
0.15 |
| chr15_81896525_81896724 | 0.13 |
Aco2 |
aconitase 2, mitochondrial |
1318 |
0.26 |
| chr2_51147047_51147231 | 0.13 |
Rnd3 |
Rho family GTPase 3 |
1955 |
0.41 |
| chr2_103485095_103485448 | 0.13 |
Cat |
catalase |
111 |
0.97 |
| chr6_125161019_125161494 | 0.13 |
Gapdh |
glyceraldehyde-3-phosphate dehydrogenase |
1271 |
0.2 |
| chr7_127026907_127027653 | 0.13 |
Maz |
MYC-associated zinc finger protein (purine-binding transcription factor) |
243 |
0.69 |
| chr15_78845338_78845568 | 0.13 |
Cdc42ep1 |
CDC42 effector protein (Rho GTPase binding) 1 |
2829 |
0.13 |
| chr5_151059334_151059485 | 0.13 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
29853 |
0.18 |
| chr11_6111840_6112016 | 0.13 |
Nudcd3 |
NudC domain containing 3 |
7481 |
0.17 |
| chr5_66679334_66679485 | 0.13 |
Uchl1 |
ubiquitin carboxy-terminal hydrolase L1 |
2517 |
0.2 |
| chr15_81815014_81815199 | 0.13 |
Tef |
thyrotroph embryonic factor |
2947 |
0.14 |
| chr8_105131845_105132023 | 0.13 |
Ces4a |
carboxylesterase 4A |
134 |
0.92 |
| chr12_76369660_76370048 | 0.13 |
Zbtb25 |
zinc finger and BTB domain containing 25 |
252 |
0.62 |
| chr19_4493105_4493260 | 0.13 |
2010003K11Rik |
RIKEN cDNA 2010003K11 gene |
5401 |
0.13 |
| chr16_22163049_22163219 | 0.13 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
115 |
0.96 |
| chr8_104926939_104927098 | 0.13 |
Ces2e |
carboxylesterase 2E |
754 |
0.48 |
| chr5_38486025_38486222 | 0.13 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
2738 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.3 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.2 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.3 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching(GO:0045829) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0018119 | peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.0 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.2 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |