Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
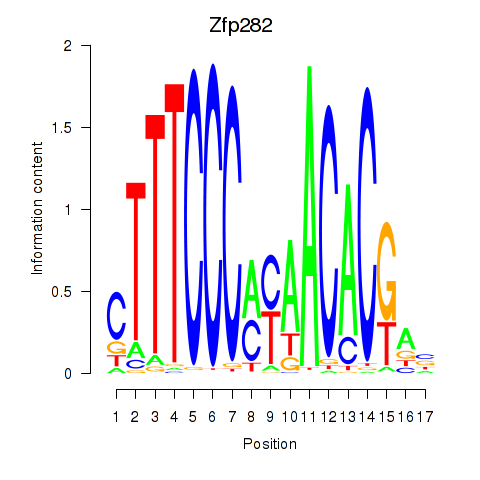
Results for Zfp282
Z-value: 1.52
Transcription factors associated with Zfp282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp282
|
ENSMUSG00000025821.9 | zinc finger protein 282 |
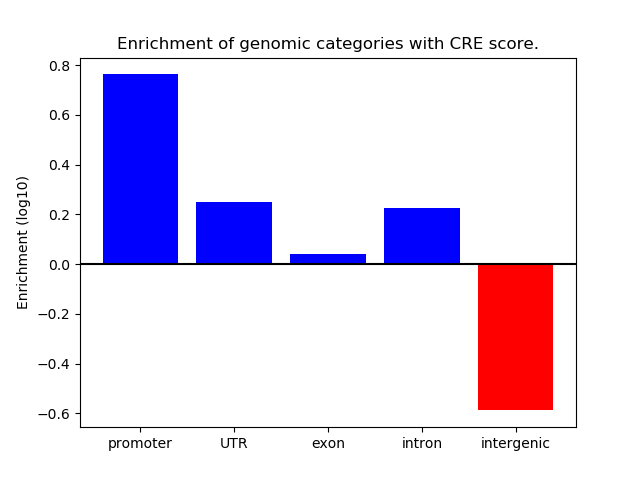
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_47877547_47877732 | Zfp282 | 17 | 0.964150 | -0.79 | 6.0e-02 | Click! |
| chr6_47877737_47877913 | Zfp282 | 169 | 0.923592 | -0.62 | 1.9e-01 | Click! |
| chr6_47879323_47879519 | Zfp282 | 1765 | 0.249621 | -0.33 | 5.2e-01 | Click! |
| chr6_47878038_47878359 | Zfp282 | 542 | 0.674152 | 0.29 | 5.7e-01 | Click! |
| chr6_47878452_47878609 | Zfp282 | 874 | 0.477761 | 0.01 | 9.8e-01 | Click! |
Activity of the Zfp282 motif across conditions
Conditions sorted by the z-value of the Zfp282 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
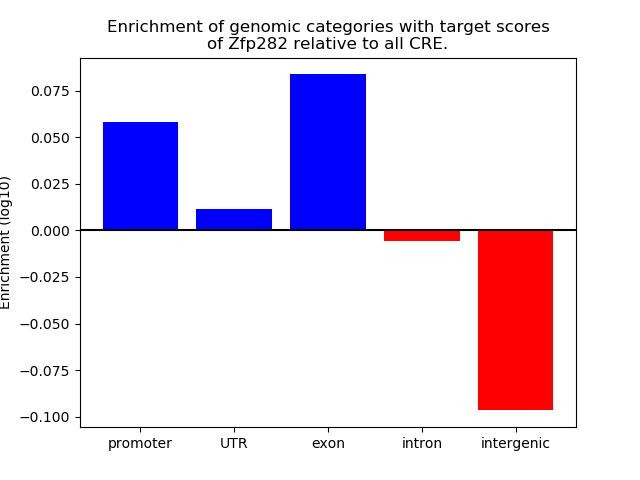
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_35866032_35866679 | 1.04 |
Ppp1r18 |
protein phosphatase 1, regulatory subunit 18 |
260 |
0.6 |
| chr2_58792449_58792713 | 0.97 |
Upp2 |
uridine phosphorylase 2 |
27256 |
0.17 |
| chr12_87443355_87443540 | 0.92 |
Alkbh1 |
alkB homolog 1, histone H2A dioxygenase |
379 |
0.44 |
| chr10_95144611_95144805 | 0.78 |
Gm29684 |
predicted gene, 29684 |
1117 |
0.44 |
| chr12_16563277_16563428 | 0.77 |
Lpin1 |
lipin 1 |
908 |
0.68 |
| chr11_120813788_120814608 | 0.76 |
Fasn |
fatty acid synthase |
1257 |
0.26 |
| chr14_120498503_120498675 | 0.70 |
Rap2a |
RAS related protein 2a |
20145 |
0.24 |
| chr8_93168995_93169226 | 0.69 |
Ces1d |
carboxylesterase 1D |
865 |
0.51 |
| chr16_24877530_24877720 | 0.67 |
Gm22672 |
predicted gene, 22672 |
6548 |
0.24 |
| chr14_68809096_68809247 | 0.67 |
Gm47256 |
predicted gene, 47256 |
65664 |
0.11 |
| chrX_85827473_85827635 | 0.64 |
Gm14762 |
predicted gene 14762 |
893 |
0.5 |
| chr11_69098275_69098686 | 0.62 |
Per1 |
period circadian clock 1 |
468 |
0.58 |
| chr14_11358016_11358174 | 0.61 |
Gm3839 |
predicted pseudogene 3839 |
1369 |
0.42 |
| chr1_75045469_75045620 | 0.60 |
Nhej1 |
non-homologous end joining factor 1 |
1095 |
0.44 |
| chr1_133348155_133348321 | 0.59 |
Ren1 |
renin 1 structural |
2272 |
0.2 |
| chr11_105905841_105906025 | 0.58 |
Tanc2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
3950 |
0.17 |
| chr13_4270977_4271134 | 0.58 |
Akr1c12 |
aldo-keto reductase family 1, member C12 |
8378 |
0.15 |
| chr12_16864896_16865104 | 0.57 |
Gm36495 |
predicted gene, 36495 |
14119 |
0.14 |
| chr4_53262351_53262525 | 0.56 |
AI427809 |
expressed sequence AI427809 |
109 |
0.96 |
| chr5_91259410_91259561 | 0.56 |
Gm19619 |
predicted gene, 19619 |
23936 |
0.19 |
| chr4_53475197_53475646 | 0.54 |
Slc44a1 |
solute carrier family 44, member 1 |
34737 |
0.17 |
| chr3_93469367_93469518 | 0.53 |
Tchhl1 |
trichohyalin-like 1 |
688 |
0.49 |
| chr8_110000217_110000533 | 0.53 |
Tat |
tyrosine aminotransferase |
9869 |
0.12 |
| chr4_137891903_137892213 | 0.52 |
Gm13012 |
predicted gene 13012 |
8121 |
0.21 |
| chr11_90371132_90371283 | 0.51 |
Hlf |
hepatic leukemia factor |
10728 |
0.26 |
| chr10_96649866_96650017 | 0.50 |
Btg1 |
BTG anti-proliferation factor 1 |
32387 |
0.16 |
| chr7_30514994_30515293 | 0.50 |
Arhgap33os |
Rho GTPase activating protein 33, opposite strand |
21 |
0.92 |
| chr10_81414192_81414360 | 0.49 |
Mir1191b |
microRNA 1191b |
2021 |
0.11 |
| chr2_156392250_156392474 | 0.49 |
2900097C17Rik |
RIKEN cDNA 2900097C17 gene |
467 |
0.64 |
| chr2_135035898_135036246 | 0.48 |
4930545L23Rik |
RIKEN cDNA 4930545L23 gene |
179544 |
0.03 |
| chr2_64095519_64096038 | 0.48 |
Fign |
fidgetin |
2210 |
0.48 |
| chr2_160364625_160364776 | 0.48 |
Mafb |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
2365 |
0.37 |
| chr4_14810853_14811073 | 0.48 |
Lrrc69 |
leucine rich repeat containing 69 |
14903 |
0.18 |
| chr6_120008206_120008357 | 0.48 |
Gm16199 |
predicted gene 16199 |
2283 |
0.27 |
| chr2_109919434_109919722 | 0.48 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
1931 |
0.32 |
| chr5_112287930_112288081 | 0.47 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
757 |
0.52 |
| chr2_92381925_92382198 | 0.47 |
1700029I15Rik |
RIKEN cDNA 1700029I15 gene |
477 |
0.66 |
| chr3_97637473_97637939 | 0.47 |
Fmo5 |
flavin containing monooxygenase 5 |
8823 |
0.13 |
| chr3_84813766_84813917 | 0.47 |
Fbxw7 |
F-box and WD-40 domain protein 7 |
1427 |
0.53 |
| chr8_88203572_88203796 | 0.46 |
Tent4b |
terminal nucleotidyltransferase 4B |
4470 |
0.18 |
| chr8_11021555_11021713 | 0.46 |
Irs2 |
insulin receptor substrate 2 |
13176 |
0.13 |
| chr14_21295488_21295639 | 0.46 |
Adk |
adenosine kinase |
22562 |
0.24 |
| chr6_138141951_138142295 | 0.45 |
Mgst1 |
microsomal glutathione S-transferase 1 |
537 |
0.85 |
| chr16_28895542_28895693 | 0.45 |
Mb21d2 |
Mab-21 domain containing 2 |
34056 |
0.2 |
| chr7_72209931_72210100 | 0.45 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
19345 |
0.23 |
| chr15_99032785_99032993 | 0.45 |
Tuba1c |
tubulin, alpha 1C |
2568 |
0.14 |
| chr13_52736115_52736276 | 0.43 |
BB123696 |
expressed sequence BB123696 |
587 |
0.84 |
| chr16_14152143_14152374 | 0.43 |
Marf1 |
meiosis regulator and mRNA stability 1 |
7009 |
0.14 |
| chr5_28061144_28061343 | 0.43 |
Gm26608 |
predicted gene, 26608 |
5784 |
0.17 |
| chr7_127800722_127801079 | 0.43 |
Hsd3b7 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
16 |
0.92 |
| chr11_16837434_16837735 | 0.43 |
Egfros |
epidermal growth factor receptor, opposite strand |
6882 |
0.22 |
| chr15_83202825_83203000 | 0.42 |
7530414M10Rik |
RIKEN cDNA 7530414M10 gene |
26467 |
0.1 |
| chr9_102981639_102981813 | 0.42 |
Slco2a1 |
solute carrier organic anion transporter family, member 2a1 |
6986 |
0.18 |
| chr18_36725667_36726187 | 0.42 |
Cd14 |
CD14 antigen |
811 |
0.38 |
| chr14_63326787_63326957 | 0.41 |
Gm5463 |
predicted gene 5463 |
2348 |
0.25 |
| chr2_27735063_27735214 | 0.41 |
Rxra |
retinoid X receptor alpha |
5063 |
0.29 |
| chr2_18031546_18031742 | 0.41 |
Gm13330 |
predicted gene 13330 |
889 |
0.42 |
| chr12_81860812_81860999 | 0.41 |
Pcnx |
pecanex homolog |
564 |
0.78 |
| chr9_23224794_23225147 | 0.40 |
Bmper |
BMP-binding endothelial regulator |
1684 |
0.55 |
| chr5_51874876_51875093 | 0.40 |
Gm42616 |
predicted gene 42616 |
4618 |
0.19 |
| chr14_88463159_88463310 | 0.40 |
Pcdh20 |
protocadherin 20 |
8112 |
0.23 |
| chr7_99062840_99062991 | 0.40 |
Gm33882 |
predicted gene, 33882 |
27317 |
0.14 |
| chr11_60782221_60782394 | 0.40 |
Smcr8 |
Smith-Magenis syndrome chromosome region, candidate 8 homolog (human) |
4782 |
0.09 |
| chr6_114866683_114866890 | 0.39 |
Vgll4 |
vestigial like family member 4 |
2029 |
0.33 |
| chr3_138261022_138261219 | 0.39 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
129 |
0.94 |
| chr1_150993778_150993941 | 0.39 |
Hmcn1 |
hemicentin 1 |
424 |
0.83 |
| chr3_51226339_51227024 | 0.39 |
Noct |
nocturnin |
2211 |
0.23 |
| chr5_34409004_34409235 | 0.39 |
Fam193a |
family with sequence homology 193, member A |
17218 |
0.13 |
| chr18_84862488_84862777 | 0.39 |
Gm16146 |
predicted gene 16146 |
3046 |
0.21 |
| chr6_119419039_119419208 | 0.38 |
Adipor2 |
adiponectin receptor 2 |
1419 |
0.44 |
| chr8_122979971_122980234 | 0.38 |
Gm24445 |
predicted gene, 24445 |
2841 |
0.19 |
| chr9_20869353_20869677 | 0.38 |
Shfl |
shiftless antiviral inhibitor of ribosomal frameshifting |
238 |
0.82 |
| chr13_9493955_9494714 | 0.38 |
Gm48871 |
predicted gene, 48871 |
48434 |
0.12 |
| chr15_74708640_74708963 | 0.38 |
4933427E11Rik |
RIKEN cDNA 4933427E11 gene |
360 |
0.59 |
| chr6_129095497_129095663 | 0.38 |
Clec2e |
C-type lectin domain family 2, member e |
5333 |
0.12 |
| chr6_54048644_54048819 | 0.38 |
Chn2 |
chimerin 2 |
8645 |
0.22 |
| chr3_88144051_88144795 | 0.37 |
Mef2d |
myocyte enhancer factor 2D |
1849 |
0.2 |
| chr11_43170710_43170884 | 0.37 |
Atp10b |
ATPase, class V, type 10B |
15126 |
0.2 |
| chr10_87896571_87896873 | 0.37 |
Igf1os |
insulin-like growth factor 1, opposite strand |
33341 |
0.15 |
| chr4_63084799_63084953 | 0.37 |
Zfp618 |
zinc finger protein 618 |
35524 |
0.16 |
| chr4_62703120_62703271 | 0.37 |
Rgs3 |
regulator of G-protein signaling 3 |
2500 |
0.23 |
| chr6_6149940_6150263 | 0.36 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
23850 |
0.23 |
| chr14_60980883_60981065 | 0.36 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
5089 |
0.27 |
| chr9_21227746_21227897 | 0.36 |
Gm16754 |
predicted gene, 16754 |
2418 |
0.14 |
| chr5_124424178_124424509 | 0.36 |
Gm37415 |
predicted gene, 37415 |
354 |
0.67 |
| chr10_67189121_67189327 | 0.36 |
Jmjd1c |
jumonji domain containing 1C |
3471 |
0.26 |
| chr9_46067501_46067769 | 0.36 |
Sik3 |
SIK family kinase 3 |
54635 |
0.09 |
| chr10_118455975_118456145 | 0.35 |
Gm49751 |
predicted gene, 49751 |
14641 |
0.16 |
| chr16_11066434_11066585 | 0.35 |
Snn |
stannin |
179 |
0.68 |
| chr5_32458502_32458674 | 0.35 |
Ppp1cb |
protein phosphatase 1 catalytic subunit beta |
255 |
0.83 |
| chr4_133162666_133162838 | 0.35 |
Wasf2 |
WAS protein family, member 2 |
13249 |
0.14 |
| chr11_101416742_101416924 | 0.35 |
Aarsd1 |
alanyl-tRNA synthetase domain containing 1 |
549 |
0.46 |
| chr10_20148225_20148689 | 0.34 |
Map7 |
microtubule-associated protein 7 |
14 |
0.98 |
| chr2_53190893_53191069 | 0.34 |
Prpf40a |
pre-mRNA processing factor 40A |
191 |
0.87 |
| chr2_34774806_34775255 | 0.34 |
Hspa5 |
heat shock protein 5 |
183 |
0.92 |
| chr2_155070325_155070499 | 0.34 |
Gm45609 |
predicted gene 45609 |
3769 |
0.16 |
| chr8_46512530_46512928 | 0.34 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
14057 |
0.15 |
| chr14_35144080_35144240 | 0.34 |
Gm49034 |
predicted gene, 49034 |
75278 |
0.12 |
| chr16_46100408_46100565 | 0.34 |
Gm50487 |
predicted gene, 50487 |
14011 |
0.18 |
| chr14_21090695_21090906 | 0.34 |
Adk |
adenosine kinase |
14648 |
0.21 |
| chr15_103273039_103273393 | 0.33 |
Copz1 |
coatomer protein complex, subunit zeta 1 |
304 |
0.8 |
| chr4_86897207_86897408 | 0.33 |
Acer2 |
alkaline ceramidase 2 |
22893 |
0.19 |
| chr10_80684012_80684199 | 0.33 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
5993 |
0.09 |
| chr9_57262499_57262900 | 0.33 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
87 |
0.96 |
| chr14_52196558_52196748 | 0.33 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
763 |
0.35 |
| chr8_20262284_20262575 | 0.33 |
6820431F20Rik |
RIKEN cDNA 6820431F20 gene |
20837 |
0.21 |
| chr10_20153471_20153802 | 0.33 |
Map7 |
microtubule-associated protein 7 |
4638 |
0.2 |
| chr17_32272780_32272931 | 0.33 |
Brd4 |
bromodomain containing 4 |
490 |
0.72 |
| chr5_66034454_66034632 | 0.33 |
Rbm47 |
RNA binding motif protein 47 |
20009 |
0.11 |
| chr4_45403274_45403659 | 0.32 |
Slc25a51 |
solute carrier family 25, member 51 |
1400 |
0.35 |
| chr7_115674458_115674622 | 0.32 |
Sox6 |
SRY (sex determining region Y)-box 6 |
12175 |
0.3 |
| chr7_90426036_90426299 | 0.32 |
Ccdc89 |
coiled-coil domain containing 89 |
410 |
0.78 |
| chr6_115717866_115718504 | 0.32 |
Gm24008 |
predicted gene, 24008 |
5647 |
0.12 |
| chr4_135789148_135789435 | 0.32 |
Myom3 |
myomesin family, member 3 |
8899 |
0.14 |
| chr1_70928023_70928290 | 0.31 |
Gm16236 |
predicted gene 16236 |
111874 |
0.07 |
| chr18_69346822_69346973 | 0.31 |
Tcf4 |
transcription factor 4 |
402 |
0.9 |
| chr2_58775472_58775623 | 0.31 |
Upp2 |
uridine phosphorylase 2 |
10222 |
0.21 |
| chr13_95828839_95829073 | 0.31 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
62801 |
0.1 |
| chr3_14853846_14854027 | 0.31 |
Car3 |
carbonic anhydrase 3 |
9576 |
0.18 |
| chr4_14865092_14865411 | 0.31 |
Pip4p2 |
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
1175 |
0.52 |
| chr19_4587227_4587438 | 0.31 |
Pcx |
pyruvate carboxylase |
7014 |
0.13 |
| chr2_69862179_69862453 | 0.31 |
Ssb |
Sjogren syndrome antigen B |
630 |
0.59 |
| chr10_19552959_19553256 | 0.31 |
Gm48563 |
predicted gene, 48563 |
30457 |
0.15 |
| chr10_75782918_75783155 | 0.31 |
Gstt3 |
glutathione S-transferase, theta 3 |
1622 |
0.19 |
| chr7_114778861_114779234 | 0.30 |
Insc |
INSC spindle orientation adaptor protein |
7726 |
0.19 |
| chr3_52267357_52267508 | 0.30 |
Foxo1 |
forkhead box O1 |
904 |
0.44 |
| chr17_80676724_80676977 | 0.30 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
25700 |
0.2 |
| chr4_122955991_122956232 | 0.30 |
Mfsd2a |
major facilitator superfamily domain containing 2A |
3879 |
0.16 |
| chr13_59536214_59536381 | 0.30 |
Agtpbp1 |
ATP/GTP binding protein 1 |
2225 |
0.26 |
| chr4_155234884_155235204 | 0.30 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
12452 |
0.15 |
| chr18_51125971_51126159 | 0.30 |
Prr16 |
proline rich 16 |
8327 |
0.31 |
| chr6_116895920_116896071 | 0.29 |
Gm43926 |
predicted gene, 43926 |
57524 |
0.12 |
| chr8_56564027_56564192 | 0.29 |
Gm25992 |
predicted gene, 25992 |
2808 |
0.19 |
| chr6_29694571_29694929 | 0.29 |
Tspan33 |
tetraspanin 33 |
516 |
0.77 |
| chr5_124421900_124422062 | 0.29 |
Sbno1 |
strawberry notch 1 |
2094 |
0.17 |
| chr6_70843849_70844027 | 0.29 |
Eif2ak3 |
eukaryotic translation initiation factor 2 alpha kinase 3 |
577 |
0.7 |
| chr12_104342354_104343298 | 0.29 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
4340 |
0.13 |
| chr6_87277564_87277945 | 0.29 |
Gm44418 |
predicted gene, 44418 |
9 |
0.96 |
| chr17_24632818_24632990 | 0.29 |
Nthl1 |
nth (endonuclease III)-like 1 (E.coli) |
185 |
0.57 |
| chr13_43530666_43530856 | 0.28 |
Gm32939 |
predicted gene, 32939 |
541 |
0.71 |
| chr2_134554640_134554803 | 0.28 |
Hao1 |
hydroxyacid oxidase 1, liver |
353 |
0.93 |
| chr8_114148783_114148961 | 0.28 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
15230 |
0.26 |
| chr17_46680513_46680702 | 0.28 |
Mea1 |
male enhanced antigen 1 |
88 |
0.62 |
| chr13_37434601_37434752 | 0.28 |
Gm4035 |
predicted gene 4035 |
473 |
0.7 |
| chr15_39816605_39817118 | 0.28 |
Gm16291 |
predicted gene 16291 |
19816 |
0.18 |
| chr5_99284196_99284371 | 0.28 |
Gm35394 |
predicted gene, 35394 |
10188 |
0.24 |
| chr1_58734421_58734701 | 0.28 |
Cflar |
CASP8 and FADD-like apoptosis regulator |
4009 |
0.16 |
| chr11_32224531_32225077 | 0.28 |
Mpg |
N-methylpurine-DNA glycosylase |
1701 |
0.21 |
| chr4_53133117_53133354 | 0.28 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
26660 |
0.18 |
| chr13_9765507_9765695 | 0.27 |
Zmynd11 |
zinc finger, MYND domain containing 11 |
271 |
0.89 |
| chr10_54058863_54059129 | 0.27 |
Gm47917 |
predicted gene, 47917 |
4815 |
0.22 |
| chr1_92667223_92667402 | 0.27 |
Otos |
otospiralin |
18471 |
0.1 |
| chr17_32492940_32493118 | 0.27 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
871 |
0.5 |
| chr4_120816433_120816604 | 0.27 |
Nfyc |
nuclear transcription factor-Y gamma |
806 |
0.51 |
| chr12_3890925_3891725 | 0.27 |
Dnmt3a |
DNA methyltransferase 3A |
403 |
0.83 |
| chr2_84996201_84996436 | 0.27 |
Prg3 |
proteoglycan 3 |
8103 |
0.12 |
| chr3_130214224_130214407 | 0.27 |
Gm22682 |
predicted gene, 22682 |
8850 |
0.21 |
| chrX_23710209_23710816 | 0.27 |
Wdr44 |
WD repeat domain 44 |
17353 |
0.28 |
| chr9_78513509_78513701 | 0.27 |
Gm47430 |
predicted gene, 47430 |
6941 |
0.13 |
| chr7_27329424_27329588 | 0.27 |
Ltbp4 |
latent transforming growth factor beta binding protein 4 |
4107 |
0.13 |
| chr19_29978982_29979139 | 0.27 |
Gm26046 |
predicted gene, 26046 |
7039 |
0.18 |
| chr7_34230685_34230836 | 0.27 |
Gpi1 |
glucose-6-phosphate isomerase 1 |
424 |
0.67 |
| chr2_165791815_165791993 | 0.27 |
Eya2 |
EYA transcriptional coactivator and phosphatase 2 |
22718 |
0.17 |
| chr1_31094512_31094669 | 0.26 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
1864 |
0.29 |
| chr16_4321605_4321756 | 0.26 |
Gm6142 |
predicted pseudogene 6142 |
1000 |
0.59 |
| chr1_88044569_88044899 | 0.26 |
AC087801.1 |
UDP glycosyltransferase 1 family (Ytg1) pseudogene |
7134 |
0.08 |
| chr2_90494801_90494975 | 0.26 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
15638 |
0.17 |
| chr19_29668488_29668647 | 0.26 |
Ermp1 |
endoplasmic reticulum metallopeptidase 1 |
20152 |
0.17 |
| chr5_123001764_123001915 | 0.26 |
Gm44574 |
predicted gene 44574 |
11035 |
0.09 |
| chr3_67430526_67430935 | 0.26 |
Gfm1 |
G elongation factor, mitochondrial 1 |
614 |
0.7 |
| chr2_64971609_64972024 | 0.26 |
Grb14 |
growth factor receptor bound protein 14 |
3930 |
0.34 |
| chr2_63861256_63861458 | 0.26 |
Fign |
fidgetin |
236631 |
0.02 |
| chrX_76137735_76137886 | 0.26 |
Gm14710 |
predicted gene 14710 |
24795 |
0.17 |
| chr5_123076789_123076960 | 0.26 |
Tmem120b |
transmembrane protein 120B |
504 |
0.62 |
| chr15_59657545_59657701 | 0.25 |
Trib1 |
tribbles pseudokinase 1 |
8970 |
0.2 |
| chr13_82201076_82201227 | 0.25 |
Gm48155 |
predicted gene, 48155 |
111394 |
0.07 |
| chr2_134549597_134549818 | 0.25 |
Hao1 |
hydroxyacid oxidase 1, liver |
4600 |
0.33 |
| chr4_9825489_9825685 | 0.25 |
Gdf6 |
growth differentiation factor 6 |
18785 |
0.21 |
| chr5_145190659_145190910 | 0.25 |
Atp5j2 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
727 |
0.48 |
| chr6_141944415_141944591 | 0.25 |
Slco1a1 |
solute carrier organic anion transporter family, member 1a1 |
2307 |
0.35 |
| chr10_24951900_24952743 | 0.25 |
Gm36172 |
predicted gene, 36172 |
24702 |
0.12 |
| chr7_130989257_130989408 | 0.25 |
Htra1 |
HtrA serine peptidase 1 |
6602 |
0.21 |
| chr2_32612430_32612590 | 0.25 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
3467 |
0.1 |
| chr9_69623415_69623593 | 0.25 |
Gm47203 |
predicted gene, 47203 |
23750 |
0.21 |
| chr4_74251698_74252250 | 0.25 |
Kdm4c |
lysine (K)-specific demethylase 4C |
38 |
0.78 |
| chr14_105176440_105176633 | 0.25 |
Rbm26 |
RNA binding motif protein 26 |
324 |
0.87 |
| chr5_135957340_135957526 | 0.25 |
Ssc4d |
scavenger receptor cysteine rich family, 4 domains |
4838 |
0.13 |
| chr15_53166677_53166864 | 0.25 |
Ext1 |
exostosin glycosyltransferase 1 |
29485 |
0.26 |
| chr10_8928866_8929030 | 0.25 |
Gm48728 |
predicted gene, 48728 |
23001 |
0.15 |
| chr18_12655181_12655597 | 0.25 |
Gm41668 |
predicted gene, 41668 |
6970 |
0.15 |
| chr18_12729849_12730702 | 0.25 |
Ttc39c |
tetratricopeptide repeat domain 39C |
4430 |
0.15 |
| chr15_75314247_75314423 | 0.25 |
Gm3454 |
predicted gene 3454 |
629 |
0.56 |
| chr13_53377028_53377241 | 0.24 |
Sptlc1 |
serine palmitoyltransferase, long chain base subunit 1 |
214 |
0.94 |
| chr16_24448114_24448329 | 0.24 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
130 |
0.97 |
| chr6_29772335_29772499 | 0.24 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
3889 |
0.2 |
| chr12_72536046_72536412 | 0.24 |
Pcnx4 |
pecanex homolog 4 |
154 |
0.96 |
| chr12_41851974_41852350 | 0.24 |
Gm47371 |
predicted gene, 47371 |
23639 |
0.25 |
| chr12_54162747_54163149 | 0.24 |
Egln3 |
egl-9 family hypoxia-inducible factor 3 |
40912 |
0.13 |
| chr9_47529383_47529534 | 0.24 |
Cadm1 |
cell adhesion molecule 1 |
715 |
0.67 |
| chr17_12941947_12942098 | 0.24 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
1422 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.7 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.3 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.3 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.2 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.2 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.2 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.0 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0035912 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:2000618 | regulation of histone H4-K16 acetylation(GO:2000618) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.0 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.0 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0001798 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:1901341 | positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.0 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.0 | 0.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.0 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:1903671 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.4 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0043733 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0052622 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |