Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
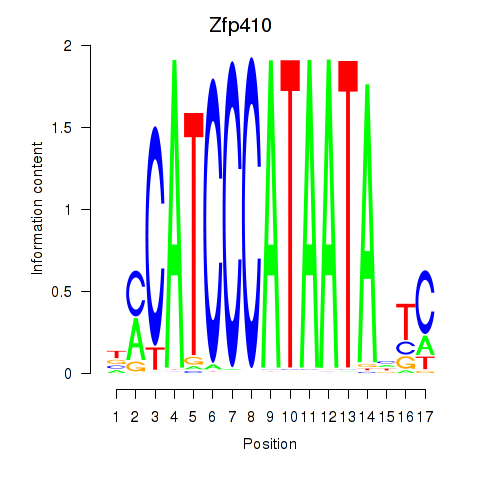
Results for Zfp410
Z-value: 0.93
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSMUSG00000042472.10 | zinc finger protein 410 |
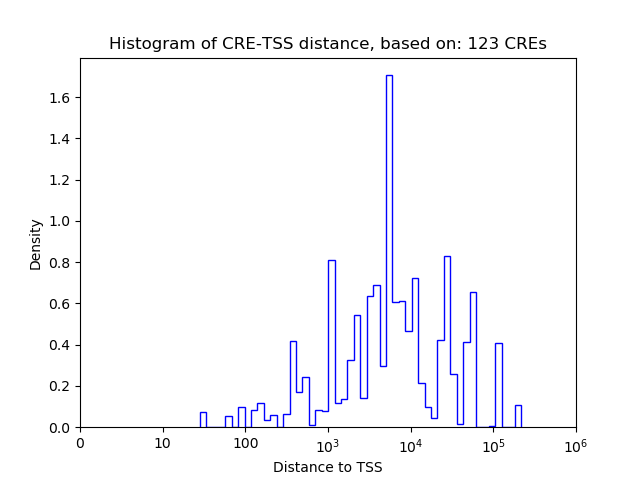
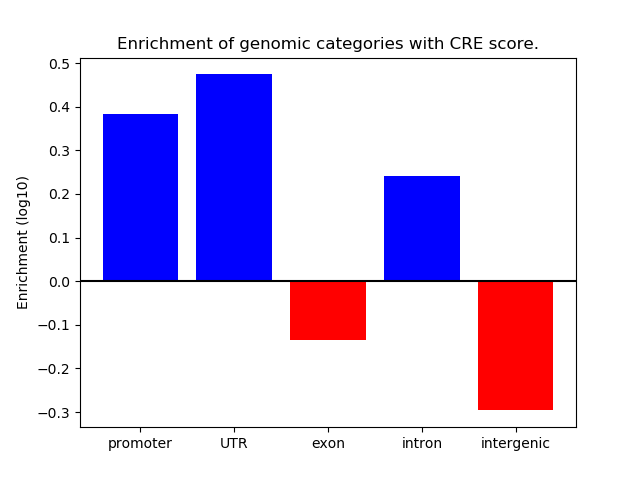
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_84317174_84317418 | Zfp410 | 151 | 0.583941 | -0.91 | 1.1e-02 | Click! |
| chr12_84316457_84316643 | Zfp410 | 302 | 0.637650 | -0.88 | 2.0e-02 | Click! |
| chr12_84317443_84317602 | Zfp410 | 377 | 0.499922 | -0.85 | 3.4e-02 | Click! |
| chr12_84316137_84316351 | Zfp410 | 608 | 0.465766 | -0.72 | 1.1e-01 | Click! |
| chr12_84319868_84320028 | Zfp410 | 2803 | 0.154894 | -0.57 | 2.3e-01 | Click! |
Activity of the Zfp410 motif across conditions
Conditions sorted by the z-value of the Zfp410 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
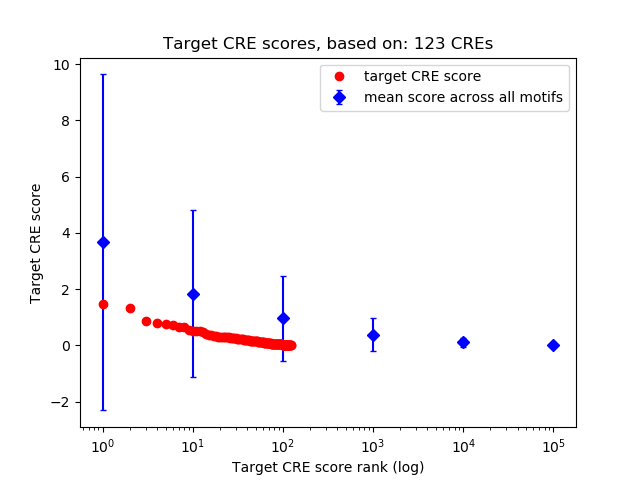
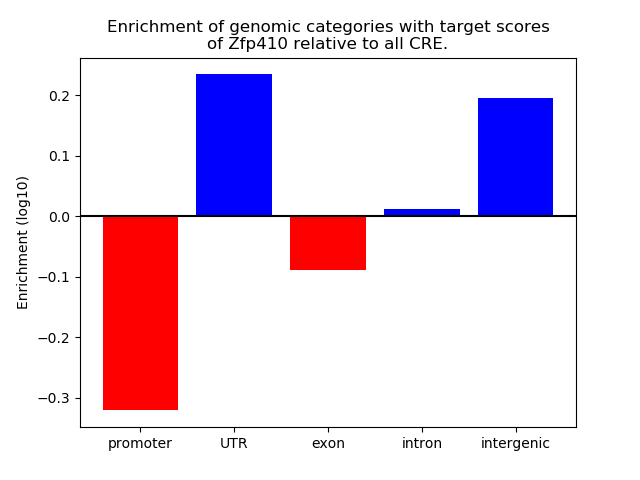
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_125088989_125089157 | 1.48 |
2010008C14Rik |
RIKEN cDNA 2010008C14 gene |
5636 |
0.07 |
| chr6_125089845_125090127 | 1.32 |
Chd4 |
chromodomain helicase DNA binding protein 4 |
5995 |
0.07 |
| chr3_31394556_31394733 | 0.85 |
Gm38025 |
predicted gene, 38025 |
53632 |
0.14 |
| chr10_78308725_78308893 | 0.80 |
AC160405.1 |
|
1190 |
0.23 |
| chr14_102952108_102952260 | 0.76 |
Kctd12 |
potassium channel tetramerisation domain containing 12 |
29367 |
0.14 |
| chr15_56280670_56280835 | 0.71 |
Hba-ps3 |
hemoglobin alpha, pseudogene 3 |
108363 |
0.07 |
| chr13_57920068_57920230 | 0.67 |
Spock1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
11817 |
0.26 |
| chr16_37603232_37603383 | 0.64 |
Gm46559 |
predicted gene, 46559 |
7117 |
0.16 |
| chr4_133015360_133015511 | 0.53 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
3231 |
0.23 |
| chr16_43298296_43298498 | 0.52 |
Gm37946 |
predicted gene, 37946 |
9884 |
0.19 |
| chr15_102103951_102104121 | 0.51 |
Tns2 |
tensin 2 |
389 |
0.76 |
| chr10_41545509_41545660 | 0.50 |
Ccdc162 |
coiled-coil domain containing 162 |
7265 |
0.12 |
| chr15_100674641_100674912 | 0.47 |
Cela1 |
chymotrypsin-like elastase family, member 1 |
4086 |
0.11 |
| chr5_146101972_146102158 | 0.39 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
22798 |
0.12 |
| chr4_118123096_118123255 | 0.36 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
11707 |
0.16 |
| chr12_3764256_3764408 | 0.35 |
Dtnbos |
dystrobrevin, beta, opposite strand |
2186 |
0.28 |
| chr14_34762111_34762276 | 0.34 |
A930038B10Rik |
RIKEN cDNA A930038B10 gene |
1966 |
0.27 |
| chr11_20381578_20381759 | 0.34 |
Gm12033 |
predicted gene 12033 |
45655 |
0.12 |
| chr14_26441433_26441601 | 0.30 |
Slmap |
sarcolemma associated protein |
1138 |
0.45 |
| chr4_60152840_60153361 | 0.30 |
Mup2 |
major urinary protein 2 |
1189 |
0.41 |
| chr8_126588974_126589213 | 0.29 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
4893 |
0.25 |
| chr7_127912019_127912170 | 0.29 |
Kat8 |
K(lysine) acetyltransferase 8 |
422 |
0.6 |
| chr19_10667308_10667470 | 0.28 |
Pga5 |
pepsinogen 5, group I |
3584 |
0.12 |
| chr16_4355769_4355950 | 0.28 |
Gm6142 |
predicted pseudogene 6142 |
35179 |
0.15 |
| chr4_121009286_121009437 | 0.28 |
Smap2 |
small ArfGAP 2 |
7886 |
0.13 |
| chr11_97180790_97180941 | 0.27 |
Kpnb1 |
karyopherin (importin) beta 1 |
7016 |
0.11 |
| chr1_85277288_85277439 | 0.27 |
Gm16026 |
predicted pseudogene 16026 |
2256 |
0.2 |
| chr2_6646229_6646380 | 0.26 |
Celf2 |
CUGBP, Elav-like family member 2 |
30124 |
0.23 |
| chr8_10948313_10948490 | 0.25 |
Gm44955 |
predicted gene 44955 |
511 |
0.66 |
| chr19_41339786_41339959 | 0.25 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
45224 |
0.15 |
| chr11_86960654_86960847 | 0.23 |
Ypel2 |
yippee like 2 |
11274 |
0.19 |
| chr9_31383704_31383866 | 0.23 |
Nfrkb |
nuclear factor related to kappa B binding protein |
2407 |
0.27 |
| chr11_90357443_90357732 | 0.22 |
Hlf |
hepatic leukemia factor |
24348 |
0.22 |
| chr8_77132343_77132494 | 0.22 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
4405 |
0.24 |
| chr2_27768705_27768856 | 0.22 |
Rxra |
retinoid X receptor alpha |
28579 |
0.21 |
| chr8_127157192_127157370 | 0.21 |
Pard3 |
par-3 family cell polarity regulator |
14123 |
0.26 |
| chr4_82384079_82384233 | 0.20 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
55254 |
0.15 |
| chr18_45574385_45574545 | 0.20 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
155 |
0.97 |
| chr12_76580294_76580728 | 0.19 |
Sptb |
spectrin beta, erythrocytic |
3575 |
0.19 |
| chr15_42892738_42892909 | 0.19 |
Angpt1 |
angiopoietin 1 |
215846 |
0.02 |
| chr5_145987383_145987573 | 0.18 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
4165 |
0.15 |
| chr6_125095352_125095503 | 0.17 |
Chd4 |
chromodomain helicase DNA binding protein 4 |
554 |
0.49 |
| chr6_125095984_125096158 | 0.17 |
Chd4 |
chromodomain helicase DNA binding protein 4 |
90 |
0.89 |
| chr4_108849580_108849741 | 0.17 |
Kti12 |
KTI12 homolog, chromatin associated |
1875 |
0.25 |
| chr18_42089247_42089538 | 0.16 |
Sh3rf2 |
SH3 domain containing ring finger 2 |
35682 |
0.15 |
| chr14_52196982_52197135 | 0.16 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
358 |
0.5 |
| chr11_53413647_53413813 | 0.15 |
Aff4 |
AF4/FMR2 family, member 4 |
3036 |
0.11 |
| chr3_135327376_135327667 | 0.15 |
Slc9b2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
2785 |
0.21 |
| chr9_78013799_78014144 | 0.15 |
Gm23609 |
predicted gene, 23609 |
127 |
0.91 |
| chr7_80018509_80018811 | 0.15 |
Zfp710 |
zinc finger protein 710 |
6154 |
0.13 |
| chr19_56378910_56379366 | 0.14 |
Nrap |
nebulin-related anchoring protein |
727 |
0.64 |
| chr14_20158546_20158697 | 0.14 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
13327 |
0.14 |
| chr18_12215421_12215611 | 0.13 |
Npc1 |
NPC intracellular cholesterol transporter 1 |
2962 |
0.2 |
| chr9_53301585_53301810 | 0.13 |
Exph5 |
exophilin 5 |
27 |
0.98 |
| chr12_102740400_102740552 | 0.12 |
Moap1 |
modulator of apoptosis 1 |
3169 |
0.1 |
| chr8_105466908_105467087 | 0.12 |
Tppp3 |
tubulin polymerization-promoting protein family member 3 |
1665 |
0.22 |
| chr4_60503881_60504482 | 0.11 |
Gm13773 |
predicted gene 13773 |
1340 |
0.3 |
| chr14_68830195_68830556 | 0.11 |
Gm47256 |
predicted gene, 47256 |
44460 |
0.17 |
| chr2_132247247_132247485 | 0.11 |
Tmem230 |
transmembrane protein 230 |
288 |
0.86 |
| chr4_142166276_142166451 | 0.10 |
Kazn |
kazrin, periplakin interacting protein |
7309 |
0.19 |
| chr6_67138304_67138472 | 0.10 |
A430010J10Rik |
RIKEN cDNA A430010J10 gene |
22711 |
0.14 |
| chr7_35131018_35131347 | 0.10 |
Gm35665 |
predicted gene, 35665 |
10267 |
0.1 |
| chr11_3134387_3135116 | 0.10 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
1256 |
0.35 |
| chr14_64949895_64950076 | 0.09 |
Ints9 |
integrator complex subunit 9 |
60 |
0.57 |
| chr3_60529673_60530018 | 0.09 |
Mbnl1 |
muscleblind like splicing factor 1 |
214 |
0.95 |
| chr3_108714394_108714793 | 0.09 |
Gpsm2 |
G-protein signalling modulator 2 (AGS3-like, C. elegans) |
2923 |
0.18 |
| chr10_68218812_68220021 | 0.09 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
59305 |
0.13 |
| chr6_124448524_124448700 | 0.09 |
Clstn3 |
calsyntenin 3 |
1560 |
0.24 |
| chr4_133701080_133701264 | 0.08 |
Mir7227 |
microRNA 7227 |
15936 |
0.12 |
| chrX_60014574_60014973 | 0.08 |
F9 |
coagulation factor IX |
15309 |
0.19 |
| chr18_20936854_20937178 | 0.08 |
Rnf125 |
ring finger protein 125 |
7609 |
0.22 |
| chr9_104391823_104391974 | 0.07 |
Gm28548 |
predicted gene 28548 |
30066 |
0.14 |
| chr6_125087229_125087503 | 0.06 |
2010008C14Rik |
RIKEN cDNA 2010008C14 gene |
3929 |
0.08 |
| chr9_108344819_108344984 | 0.06 |
Usp4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
2952 |
0.1 |
| chr14_70317262_70317413 | 0.06 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
3412 |
0.15 |
| chr19_55258606_55258757 | 0.06 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
5312 |
0.19 |
| chr7_104507554_104507721 | 0.06 |
Trim30d |
tripartite motif-containing 30D |
188 |
0.88 |
| chr2_31733558_31733887 | 0.06 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
26221 |
0.12 |
| chr7_67228989_67229173 | 0.06 |
Lysmd4 |
LysM, putative peptidoglycan-binding, domain containing 4 |
5472 |
0.17 |
| chr1_105718739_105718916 | 0.06 |
Gm37566 |
predicted gene, 37566 |
10254 |
0.16 |
| chrX_60404794_60404961 | 0.05 |
Atp11c |
ATPase, class VI, type 11C |
896 |
0.6 |
| chr2_9014159_9014316 | 0.05 |
Gm13217 |
predicted gene 13217 |
28247 |
0.25 |
| chr16_21788095_21788271 | 0.05 |
Ehhadh |
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
376 |
0.79 |
| chr12_3892588_3892788 | 0.05 |
Dnmt3a |
DNA methyltransferase 3A |
944 |
0.53 |
| chr15_81427541_81427723 | 0.05 |
Gm17025 |
predicted gene 17025 |
8444 |
0.12 |
| chr15_54943868_54944200 | 0.04 |
Gm26684 |
predicted gene, 26684 |
8036 |
0.17 |
| chr12_79969392_79969913 | 0.04 |
Gm8275 |
predicted gene 8275 |
10199 |
0.18 |
| chr13_56630053_56630394 | 0.04 |
Tgfbi |
transforming growth factor, beta induced |
990 |
0.57 |
| chr11_70835202_70835548 | 0.04 |
Rabep1 |
rabaptin, RAB GTPase binding effector protein 1 |
9403 |
0.11 |
| chrX_70475319_70475500 | 0.04 |
Tmem185a |
transmembrane protein 185A |
1733 |
0.2 |
| chr14_25716292_25716457 | 0.04 |
Zcchc24 |
zinc finger, CCHC domain containing 24 |
3418 |
0.17 |
| chr1_31094891_31095067 | 0.04 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
1475 |
0.35 |
| chr2_29030024_29030239 | 0.03 |
Cfap77 |
cilia and flagella associated protein 77 |
9229 |
0.17 |
| chr8_126776388_126776581 | 0.03 |
Gm45805 |
predicted gene 45805 |
18150 |
0.22 |
| chrX_135205150_135205310 | 0.03 |
Tceal6 |
transcription elongation factor A (SII)-like 6 |
5457 |
0.14 |
| chr14_17733474_17733665 | 0.03 |
Gm48320 |
predicted gene, 48320 |
37553 |
0.2 |
| chr4_106827627_106828287 | 0.03 |
Gm12746 |
predicted gene 12746 |
19963 |
0.15 |
| chr4_61696485_61696867 | 0.03 |
Mup-ps15 |
major urinary protein, pseudogene 15 |
2165 |
0.27 |
| chr11_86382860_86383034 | 0.03 |
Med13 |
mediator complex subunit 13 |
25345 |
0.15 |
| chr17_36030191_36030370 | 0.02 |
H2-T23 |
histocompatibility 2, T region locus 23 |
2261 |
0.1 |
| chr19_3831749_3831913 | 0.02 |
Gm19209 |
predicted gene, 19209 |
9442 |
0.1 |
| chr9_73044177_73044334 | 0.02 |
Rab27a |
RAB27A, member RAS oncogene family |
599 |
0.52 |
| chr2_28583373_28583589 | 0.02 |
Gtf3c5 |
general transcription factor IIIC, polypeptide 5 |
202 |
0.88 |
| chr5_130184977_130185128 | 0.02 |
Rabgef1 |
RAB guanine nucleotide exchange factor (GEF) 1 |
2109 |
0.18 |
| chr16_23334291_23334455 | 0.02 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
43903 |
0.12 |
| chr16_34083289_34083602 | 0.01 |
Kalrn |
kalirin, RhoGEF kinase |
12542 |
0.25 |
| chr17_26497684_26497862 | 0.01 |
Gm34455 |
predicted gene, 34455 |
5427 |
0.12 |
| chr16_44392333_44392522 | 0.01 |
Cfap44 |
cilia and flagella associated protein 44 |
2369 |
0.31 |
| chr4_129331693_129331844 | 0.01 |
Rbbp4 |
retinoblastoma binding protein 4, chromatin remodeling factor |
2876 |
0.16 |
| chr2_52577576_52577747 | 0.01 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
19094 |
0.18 |
| chr2_6128253_6128433 | 0.01 |
Proser2 |
proline and serine rich 2 |
1796 |
0.28 |
| chr12_64968165_64968316 | 0.01 |
Togaram1 |
TOG array regulator of axonemal microtubules 1 |
2184 |
0.21 |
| chr13_49179527_49179727 | 0.01 |
Ninj1 |
ninjurin 1 |
7858 |
0.18 |
| chr14_21741929_21742087 | 0.01 |
Gm15935 |
predicted gene 15935 |
419 |
0.5 |
| chr17_4611843_4612060 | 0.01 |
4930517M08Rik |
RIKEN cDNA 4930517M08 gene |
23027 |
0.24 |
| chr17_5592858_5593167 | 0.01 |
Zdhhc14 |
zinc finger, DHHC domain containing 14 |
100455 |
0.06 |
| chr13_104706600_104706768 | 0.01 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
21753 |
0.23 |
| chr2_27684119_27684319 | 0.00 |
Rxra |
retinoid X receptor alpha |
6957 |
0.27 |
| chr2_147904893_147905044 | 0.00 |
Gm25516 |
predicted gene, 25516 |
30090 |
0.2 |
| chr4_60754481_60754836 | 0.00 |
Mup-ps8 |
major urinary protein, pseudogene 8 |
2154 |
0.29 |
| chr18_42467835_42467998 | 0.00 |
Gm3631 |
predicted gene 3631 |
19172 |
0.16 |
| chr9_9114117_9114295 | 0.00 |
Gm16833 |
predicted gene, 16833 |
122082 |
0.06 |
| chr8_37423159_37423310 | 0.00 |
Gm45654 |
predicted gene 45654 |
22214 |
0.2 |
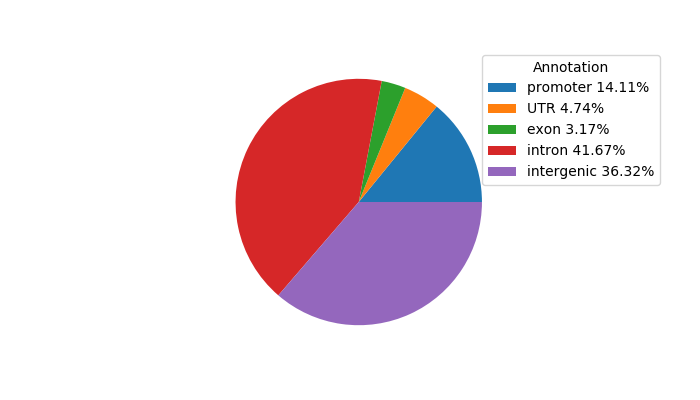
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.0 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0019178 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |