Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
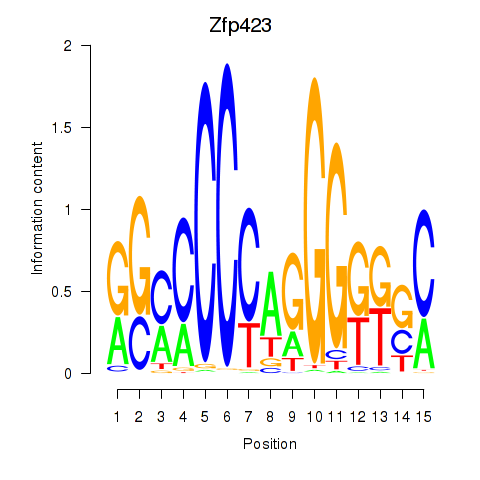
Results for Zfp423
Z-value: 1.31
Transcription factors associated with Zfp423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp423
|
ENSMUSG00000045333.9 | zinc finger protein 423 |
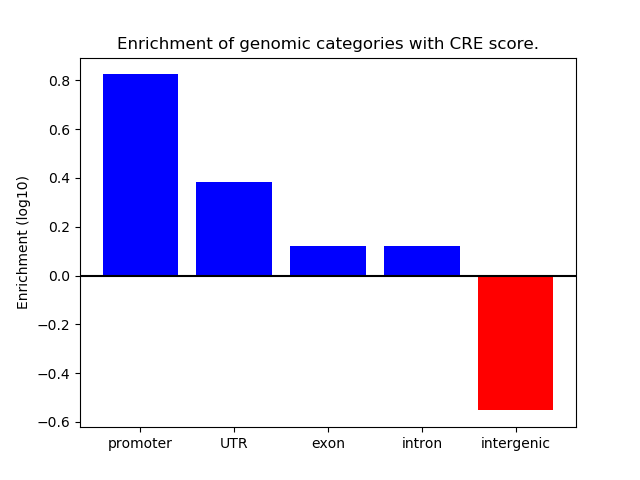
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_87958728_87958915 | Zfp423 | 774 | 0.739801 | 0.88 | 1.9e-02 | Click! |
| chr8_87961136_87961302 | Zfp423 | 1624 | 0.480154 | 0.66 | 1.6e-01 | Click! |
| chr8_87958522_87958673 | Zfp423 | 998 | 0.651721 | 0.64 | 1.7e-01 | Click! |
| chr8_87967436_87967595 | Zfp423 | 7920 | 0.261592 | -0.49 | 3.2e-01 | Click! |
| chr8_87928980_87929146 | Zfp423 | 14989 | 0.257222 | -0.47 | 3.5e-01 | Click! |
Activity of the Zfp423 motif across conditions
Conditions sorted by the z-value of the Zfp423 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
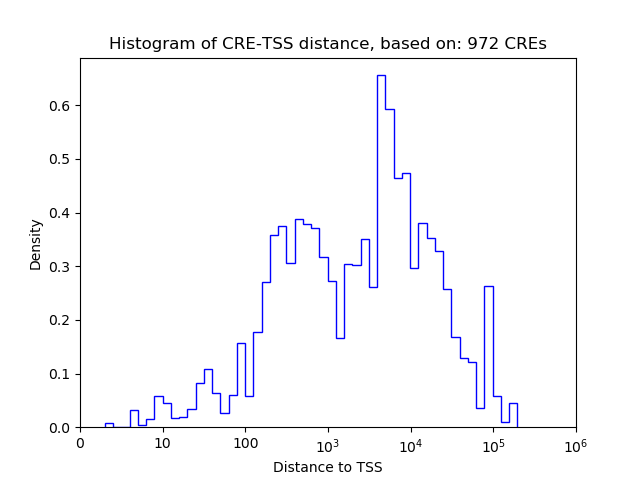
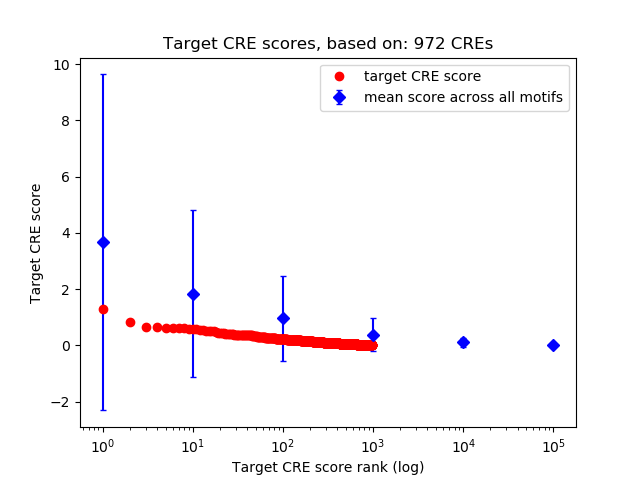
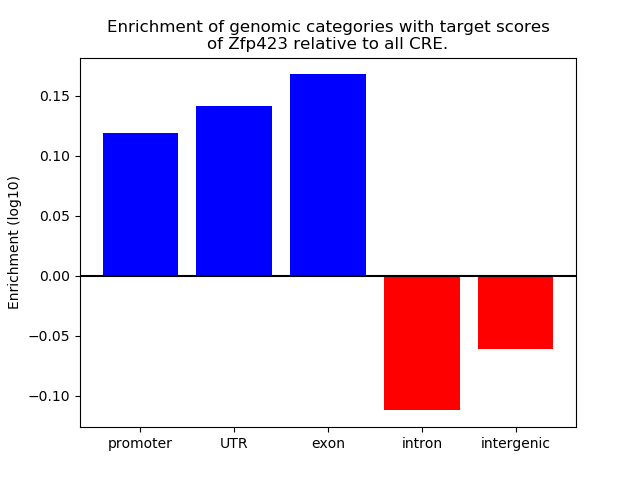
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_118232810_118233328 | 1.28 |
Gm17399 |
predicted gene, 17399 |
82838 |
0.08 |
| chr9_122056710_122056883 | 0.84 |
Gm39465 |
predicted gene, 39465 |
5333 |
0.13 |
| chr6_72607554_72607726 | 0.66 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
995 |
0.29 |
| chr7_24595623_24595793 | 0.65 |
Zfp575 |
zinc finger protein 575 |
8067 |
0.08 |
| chr12_46819464_46819646 | 0.62 |
Nova1 |
NOVA alternative splicing regulator 1 |
626 |
0.75 |
| chr1_90914963_90915133 | 0.61 |
Mlph |
melanophilin |
37 |
0.97 |
| chr9_119145447_119145657 | 0.60 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
4511 |
0.13 |
| chr14_19712945_19713096 | 0.60 |
Gm49341 |
predicted gene, 49341 |
15811 |
0.12 |
| chr17_56206282_56206453 | 0.58 |
Dpp9 |
dipeptidylpeptidase 9 |
2841 |
0.13 |
| chr6_54699097_54699279 | 0.57 |
Gm44008 |
predicted gene, 44008 |
5542 |
0.18 |
| chr4_138367286_138367444 | 0.57 |
Cda |
cytidine deaminase |
627 |
0.6 |
| chr6_121329747_121329921 | 0.56 |
Gm24855 |
predicted gene, 24855 |
4158 |
0.18 |
| chr11_75443489_75443686 | 0.54 |
Serpinf2 |
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
3996 |
0.09 |
| chr5_114146641_114146865 | 0.52 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
218 |
0.89 |
| chr6_59456281_59456457 | 0.52 |
Gprin3 |
GPRIN family member 3 |
30075 |
0.25 |
| chr17_53627390_53627541 | 0.50 |
Kat2b |
K(lysine) acetyltransferase 2B |
17232 |
0.14 |
| chr12_31074210_31074363 | 0.50 |
Fam110c |
family with sequence similarity 110, member C |
425 |
0.77 |
| chr2_6124892_6125054 | 0.46 |
Proser2 |
proline and serine rich 2 |
5166 |
0.17 |
| chr16_12924383_12924551 | 0.44 |
4930414F18Rik |
RIKEN cDNA 4930414F18 gene |
21468 |
0.21 |
| chr6_72120521_72121047 | 0.44 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr16_30266715_30266896 | 0.42 |
Cpn2 |
carboxypeptidase N, polypeptide 2 |
694 |
0.61 |
| chr13_29759437_29759751 | 0.42 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
95837 |
0.09 |
| chr7_75852547_75852704 | 0.41 |
Klhl25 |
kelch-like 25 |
4184 |
0.26 |
| chr18_38923142_38923385 | 0.41 |
Fgf1 |
fibroblast growth factor 1 |
4492 |
0.24 |
| chr2_178631647_178631798 | 0.41 |
Cdh26 |
cadherin-like 26 |
171092 |
0.03 |
| chr10_82804895_82805071 | 0.40 |
1700028I16Rik |
RIKEN cDNA 1700028I16 gene |
7140 |
0.13 |
| chr12_41247016_41247167 | 0.40 |
Gm47376 |
predicted gene, 47376 |
100999 |
0.07 |
| chr7_39967880_39968059 | 0.39 |
Gm44992 |
predicted gene 44992 |
10105 |
0.24 |
| chr11_44002192_44002372 | 0.38 |
Gm12153 |
predicted gene 12153 |
43193 |
0.18 |
| chr7_118580172_118580323 | 0.38 |
Tmc7 |
transmembrane channel-like gene family 7 |
4439 |
0.17 |
| chr18_82674862_82675025 | 0.38 |
Zfp236 |
zinc finger protein 236 |
8625 |
0.11 |
| chr17_26505091_26505576 | 0.38 |
Gm34455 |
predicted gene, 34455 |
2133 |
0.18 |
| chr17_29058763_29058940 | 0.37 |
Gm41556 |
predicted gene, 41556 |
622 |
0.52 |
| chr16_25059586_25059982 | 0.37 |
A230028O05Rik |
RIKEN cDNA A230028O05 gene |
145 |
0.98 |
| chr4_129672269_129672601 | 0.37 |
Kpna6 |
karyopherin (importin) alpha 6 |
93 |
0.94 |
| chr12_17473483_17473634 | 0.37 |
Gm36752 |
predicted gene, 36752 |
36940 |
0.13 |
| chr17_32482095_32482423 | 0.37 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
4008 |
0.16 |
| chr2_31462375_31462570 | 0.37 |
Ass1 |
argininosuccinate synthetase 1 |
7735 |
0.19 |
| chr1_93136180_93136798 | 0.36 |
Agxt |
alanine-glyoxylate aminotransferase |
921 |
0.46 |
| chr1_172141858_172142009 | 0.36 |
Gm10171 |
predicted gene 10171 |
2968 |
0.14 |
| chr7_113210463_113210620 | 0.36 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
2984 |
0.29 |
| chr11_79992533_79993138 | 0.35 |
Suz12 |
SUZ12 polycomb repressive complex 2 subunit |
271 |
0.91 |
| chr15_6626042_6626459 | 0.35 |
Fyb |
FYN binding protein |
6656 |
0.2 |
| chr7_25317114_25317265 | 0.35 |
Megf8 |
multiple EGF-like-domains 8 |
25 |
0.95 |
| chr4_135961065_135961216 | 0.35 |
Hmgcl |
3-hydroxy-3-methylglutaryl-Coenzyme A lyase |
808 |
0.4 |
| chr7_19748516_19748701 | 0.34 |
Nectin2 |
nectin cell adhesion molecule 2 |
925 |
0.32 |
| chr15_83857228_83857398 | 0.33 |
Mpped1 |
metallophosphoesterase domain containing 1 |
21273 |
0.19 |
| chr11_63891944_63892129 | 0.33 |
Hs3st3b1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
30254 |
0.17 |
| chr9_20868532_20868915 | 0.32 |
Shfl |
shiftless antiviral inhibitor of ribosomal frameshifting |
43 |
0.94 |
| chr2_94406371_94406538 | 0.32 |
Ttc17 |
tetratricopeptide repeat domain 17 |
225 |
0.58 |
| chr15_85935281_85935435 | 0.31 |
Celsr1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
12936 |
0.17 |
| chr18_31642183_31642334 | 0.31 |
Sap130 |
Sin3A associated protein |
7079 |
0.18 |
| chr12_103148529_103148680 | 0.31 |
Unc79 |
unc-79 homolog |
14063 |
0.2 |
| chr19_46659862_46660079 | 0.31 |
Gm36602 |
predicted gene, 36602 |
6156 |
0.13 |
| chr3_41491371_41491522 | 0.30 |
Platr4 |
pluripotency associated transcript 4 |
1744 |
0.27 |
| chr7_143053595_143053746 | 0.30 |
R74862 |
expressed sequence R74862 |
7 |
0.86 |
| chr12_17852257_17852416 | 0.30 |
Gm49326 |
predicted gene, 49326 |
29861 |
0.12 |
| chrX_8206941_8207115 | 0.29 |
Porcn |
porcupine O-acyltransferase |
503 |
0.68 |
| chr19_38124006_38124436 | 0.29 |
Rbp4 |
retinol binding protein 4, plasma |
504 |
0.74 |
| chr15_39870319_39870474 | 0.28 |
Dpys |
dihydropyrimidinase |
12926 |
0.21 |
| chr6_39241464_39242040 | 0.28 |
Gm43479 |
predicted gene 43479 |
1638 |
0.32 |
| chr10_75067635_75067786 | 0.28 |
Bcr |
BCR activator of RhoGEF and GTPase |
7118 |
0.18 |
| chr5_30423662_30423825 | 0.28 |
Otof |
otoferlin |
15682 |
0.15 |
| chr17_56111930_56112088 | 0.28 |
Plin4 |
perilipin 4 |
2206 |
0.14 |
| chr10_77021731_77021886 | 0.28 |
Gm3137 |
predicted gene, 3137 |
7767 |
0.12 |
| chr4_133633685_133634017 | 0.27 |
Zdhhc18 |
zinc finger, DHHC domain containing 18 |
382 |
0.77 |
| chr17_8801315_8801513 | 0.27 |
Pde10a |
phosphodiesterase 10A |
279 |
0.94 |
| chr7_98119476_98119911 | 0.27 |
Myo7a |
myosin VIIA |
169 |
0.95 |
| chr11_69802912_69803071 | 0.27 |
Fgf11 |
fibroblast growth factor 11 |
1134 |
0.19 |
| chr9_104042276_104042447 | 0.27 |
Gm28305 |
predicted gene 28305 |
1699 |
0.18 |
| chr14_17753703_17753909 | 0.26 |
Gm48320 |
predicted gene, 48320 |
17316 |
0.27 |
| chr4_126115280_126115448 | 0.26 |
Stk40 |
serine/threonine kinase 40 |
2908 |
0.15 |
| chr4_138348053_138348212 | 0.25 |
Cda |
cytidine deaminase |
4483 |
0.13 |
| chr1_53296592_53296800 | 0.25 |
Pms1 |
PMS1 homolog 1, mismatch repair system component |
271 |
0.46 |
| chr17_29073166_29073338 | 0.25 |
Trp53cor1 |
tumor protein p53 pathway corepressor 1 |
347 |
0.75 |
| chr8_121620520_121620873 | 0.25 |
Zcchc14 |
zinc finger, CCHC domain containing 14 |
7914 |
0.12 |
| chr12_111759014_111759441 | 0.25 |
Klc1 |
kinesin light chain 1 |
214 |
0.89 |
| chr17_34742637_34742870 | 0.25 |
C4b |
complement component 4B (Chido blood group) |
1129 |
0.24 |
| chr3_121531769_121531936 | 0.25 |
A530020G20Rik |
RIKEN cDNA A530020G20 gene |
173 |
0.82 |
| chr3_104780962_104781128 | 0.24 |
Ppm1j |
protein phosphatase 1J |
11 |
0.92 |
| chr10_78482196_78482378 | 0.24 |
Gm30232 |
predicted gene, 30232 |
1308 |
0.21 |
| chr10_80448589_80448759 | 0.24 |
Tcf3 |
transcription factor 3 |
15027 |
0.08 |
| chr2_154398565_154398783 | 0.24 |
Snta1 |
syntrophin, acidic 1 |
9306 |
0.16 |
| chr4_144950062_144950271 | 0.23 |
Gm38074 |
predicted gene, 38074 |
8682 |
0.2 |
| chr6_137738154_137738336 | 0.23 |
Strap |
serine/threonine kinase receptor associated protein |
3167 |
0.29 |
| chr17_28233099_28233728 | 0.23 |
Ppard |
peroxisome proliferator activator receptor delta |
606 |
0.6 |
| chr14_100221834_100222037 | 0.23 |
Gm16260 |
predicted gene 16260 |
2449 |
0.3 |
| chr11_75165433_75165593 | 0.23 |
Hic1 |
hypermethylated in cancer 1 |
2633 |
0.13 |
| chr3_95160050_95160201 | 0.23 |
Sema6c |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
332 |
0.72 |
| chr17_12931878_12932047 | 0.22 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
4280 |
0.09 |
| chr4_108780280_108780466 | 0.22 |
Zfyve9 |
zinc finger, FYVE domain containing 9 |
130 |
0.95 |
| chr9_21425419_21426009 | 0.22 |
Dnm2 |
dynamin 2 |
470 |
0.68 |
| chr6_124814399_124814561 | 0.22 |
Tpi1 |
triosephosphate isomerase 1 |
184 |
0.84 |
| chr6_90565247_90565487 | 0.22 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
2339 |
0.23 |
| chr11_62560745_62560932 | 0.22 |
Gm12280 |
predicted gene 12280 |
5545 |
0.09 |
| chr4_141546149_141546719 | 0.22 |
B330016D10Rik |
RIKEN cDNA B330016D10 gene |
245 |
0.89 |
| chr13_73505821_73505982 | 0.22 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
3040 |
0.27 |
| chr4_156093629_156093788 | 0.22 |
9430015G10Rik |
RIKEN cDNA 9430015G10 gene |
16274 |
0.08 |
| chr10_127612734_127612929 | 0.22 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
8091 |
0.1 |
| chr4_45527993_45528163 | 0.22 |
Shb |
src homology 2 domain-containing transforming protein B |
2252 |
0.24 |
| chr2_152781176_152781342 | 0.22 |
Gm23802 |
predicted gene, 23802 |
8370 |
0.12 |
| chr16_13205450_13205719 | 0.21 |
Mrtfb |
myocardin related transcription factor B |
50897 |
0.15 |
| chr11_24049318_24049496 | 0.21 |
A830031A19Rik |
RIKEN cDNA A830031A19 gene |
25647 |
0.13 |
| chr4_129928537_129928701 | 0.21 |
Spocd1 |
SPOC domain containing 1 |
630 |
0.61 |
| chr11_69580921_69581079 | 0.21 |
Trp53 |
transformation related protein 53 |
595 |
0.37 |
| chr11_97803969_97804142 | 0.21 |
Lasp1 |
LIM and SH3 protein 1 |
1137 |
0.28 |
| chrX_12045986_12046173 | 0.21 |
Bcor |
BCL6 interacting corepressor |
34474 |
0.19 |
| chr13_92530147_92530703 | 0.21 |
Zfyve16 |
zinc finger, FYVE domain containing 16 |
443 |
0.84 |
| chr7_101300105_101300356 | 0.21 |
Atg16l2 |
autophagy related 16-like 2 (S. cerevisiae) |
1797 |
0.23 |
| chr12_81155440_81155704 | 0.21 |
Mir3067 |
microRNA 3067 |
10579 |
0.22 |
| chr11_120933043_120933238 | 0.21 |
Ccdc57 |
coiled-coil domain containing 57 |
268 |
0.86 |
| chr4_150725753_150726057 | 0.21 |
Gm16079 |
predicted gene 16079 |
47113 |
0.13 |
| chr6_72097199_72097385 | 0.21 |
St3gal5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
300 |
0.84 |
| chr2_70538432_70538586 | 0.20 |
Gad1 |
glutamate decarboxylase 1 |
14563 |
0.12 |
| chr4_133257569_133257750 | 0.20 |
Sytl1 |
synaptotagmin-like 1 |
5393 |
0.12 |
| chr8_85840465_85840659 | 0.20 |
Itfg1 |
integrin alpha FG-GAP repeat containing 1 |
359 |
0.51 |
| chr8_11308621_11308772 | 0.20 |
Col4a1 |
collagen, type IV, alpha 1 |
3993 |
0.18 |
| chr9_63184215_63184671 | 0.20 |
Skor1 |
SKI family transcriptional corepressor 1 |
35482 |
0.15 |
| chr11_94171545_94171712 | 0.20 |
B230206L02Rik |
RIKEN cDNA B230206L02 gene |
35968 |
0.13 |
| chr1_184870102_184870490 | 0.20 |
C130074G19Rik |
RIKEN cDNA C130074G19 gene |
12922 |
0.15 |
| chr11_101667268_101667443 | 0.20 |
Arl4d |
ADP-ribosylation factor-like 4D |
1814 |
0.19 |
| chr2_109918614_109919408 | 0.20 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
1364 |
0.42 |
| chr7_44604134_44604335 | 0.20 |
Kcnc3 |
potassium voltage gated channel, Shaw-related subfamily, member 3 |
8141 |
0.07 |
| chr10_81192185_81192336 | 0.20 |
Dapk3 |
death-associated protein kinase 3 |
704 |
0.37 |
| chr11_78164488_78164639 | 0.20 |
Traf4 |
TNF receptor associated factor 4 |
953 |
0.27 |
| chr17_36231281_36231449 | 0.20 |
Gm18604 |
predicted gene, 18604 |
1896 |
0.09 |
| chr12_110207275_110207455 | 0.20 |
Gm34785 |
predicted gene, 34785 |
19304 |
0.11 |
| chr4_109670382_109670535 | 0.20 |
Cdkn2c |
cyclin dependent kinase inhibitor 2C |
3269 |
0.2 |
| chr9_106818557_106818729 | 0.19 |
Dcaf1 |
DDB1 and CUL4 associated factor 1 |
3231 |
0.17 |
| chrX_8182990_8183171 | 0.19 |
Ebp |
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
4094 |
0.12 |
| chr10_59769174_59769339 | 0.19 |
Micu1 |
mitochondrial calcium uptake 1 |
6843 |
0.17 |
| chr6_124414593_124414746 | 0.19 |
Pex5 |
peroxisomal biogenesis factor 5 |
194 |
0.9 |
| chr12_80325192_80325380 | 0.19 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
51406 |
0.09 |
| chr3_107631893_107632246 | 0.19 |
Strip1 |
striatin interacting protein 1 |
375 |
0.67 |
| chr12_17691531_17691833 | 0.19 |
Hpcal1 |
hippocalcin-like 1 |
826 |
0.68 |
| chr19_59367680_59367885 | 0.19 |
Pdzd8 |
PDZ domain containing 8 |
22002 |
0.13 |
| chr7_123466413_123466584 | 0.19 |
Aqp8 |
aquaporin 8 |
2747 |
0.27 |
| chr11_119089905_119090309 | 0.19 |
Cbx4 |
chromobox 4 |
3886 |
0.17 |
| chr5_24652159_24652333 | 0.19 |
1700022A21Rik |
RIKEN cDNA 1700022A21 gene |
4562 |
0.13 |
| chr12_8591645_8591809 | 0.19 |
Slc7a15 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
7339 |
0.21 |
| chr12_3926230_3926408 | 0.19 |
Gm9088 |
predicted gene 9088 |
5721 |
0.16 |
| chr11_120973422_120973891 | 0.19 |
Csnk1d |
casein kinase 1, delta |
2031 |
0.18 |
| chr18_60726300_60726451 | 0.19 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
1188 |
0.43 |
| chr12_111356343_111356494 | 0.18 |
Cdc42bpb |
CDC42 binding protein kinase beta |
21201 |
0.14 |
| chr5_143112262_143112625 | 0.18 |
Rnf216 |
ring finger protein 216 |
537 |
0.66 |
| chr17_34824182_34824366 | 0.18 |
C4a |
complement component 4A (Rodgers blood group) |
810 |
0.27 |
| chr16_38294532_38294704 | 0.18 |
Nr1i2 |
nuclear receptor subfamily 1, group I, member 2 |
206 |
0.92 |
| chr9_48836587_48836813 | 0.18 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
478 |
0.85 |
| chr14_55899852_55900071 | 0.18 |
Sdr39u1 |
short chain dehydrogenase/reductase family 39U, member 1 |
19 |
0.95 |
| chr5_103655342_103655532 | 0.18 |
1700016H13Rik |
RIKEN cDNA 1700016H13 gene |
286 |
0.86 |
| chr9_62386066_62386269 | 0.18 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
12728 |
0.2 |
| chr18_67279022_67279221 | 0.18 |
Impa2 |
inositol (myo)-1(or 4)-monophosphatase 2 |
10065 |
0.15 |
| chr2_151926945_151927109 | 0.17 |
Angpt4 |
angiopoietin 4 |
15817 |
0.12 |
| chr3_90231225_90231401 | 0.17 |
Jtb |
jumping translocation breakpoint |
284 |
0.78 |
| chr3_98294775_98294926 | 0.17 |
Gm43189 |
predicted gene 43189 |
7862 |
0.15 |
| chr10_80683730_80683904 | 0.17 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
5705 |
0.09 |
| chr17_26504366_26504899 | 0.17 |
Gm34455 |
predicted gene, 34455 |
1432 |
0.26 |
| chr9_96966105_96966535 | 0.17 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
5655 |
0.18 |
| chr9_21239006_21239167 | 0.17 |
Keap1 |
kelch-like ECH-associated protein 1 |
7 |
0.95 |
| chr1_190173088_190173263 | 0.17 |
Prox1 |
prospero homeobox 1 |
2461 |
0.25 |
| chr9_85328031_85328205 | 0.17 |
Tent5a |
terminal nucleotidyltransferase 5A |
770 |
0.59 |
| chr18_32252120_32252375 | 0.17 |
Ercc3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
5022 |
0.23 |
| chr17_5295996_5296161 | 0.17 |
Gm29050 |
predicted gene 29050 |
92685 |
0.08 |
| chr19_47338199_47338378 | 0.17 |
Sh3pxd2a |
SH3 and PX domains 2A |
23537 |
0.17 |
| chr2_151943059_151943241 | 0.17 |
Gm14154 |
predicted gene 14154 |
7491 |
0.14 |
| chr3_36483169_36483342 | 0.17 |
1810062G17Rik |
RIKEN cDNA 1810062G17 gene |
7318 |
0.12 |
| chr1_4785233_4785557 | 0.16 |
Mrpl15 |
mitochondrial ribosomal protein L15 |
297 |
0.83 |
| chr4_8754940_8755164 | 0.16 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
3614 |
0.35 |
| chr9_103091015_103091286 | 0.16 |
Gm47468 |
predicted gene, 47468 |
5613 |
0.17 |
| chr19_28834565_28834716 | 0.16 |
Slc1a1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
409 |
0.82 |
| chr11_11942496_11942647 | 0.16 |
Grb10 |
growth factor receptor bound protein 10 |
3759 |
0.24 |
| chr4_49509287_49509438 | 0.16 |
Baat |
bile acid-Coenzyme A: amino acid N-acyltransferase |
2805 |
0.17 |
| chr17_26702508_26702992 | 0.16 |
Crebrf |
CREB3 regulatory factor |
12900 |
0.14 |
| chr18_32414906_32415059 | 0.16 |
Bin1 |
bridging integrator 1 |
7348 |
0.18 |
| chr11_33398787_33398953 | 0.16 |
Anp32-ps |
acidic (leucine-rich) nuclear phosphoprotein 32 family, pseudogene |
3604 |
0.26 |
| chr12_113141793_113142049 | 0.16 |
Crip2 |
cysteine rich protein 2 |
887 |
0.41 |
| chr2_118487263_118487442 | 0.16 |
Gm13983 |
predicted gene 13983 |
3175 |
0.2 |
| chr1_190117117_190117268 | 0.16 |
Gm28172 |
predicted gene 28172 |
51478 |
0.13 |
| chr8_119574627_119574804 | 0.15 |
Hsdl1 |
hydroxysteroid dehydrogenase like 1 |
460 |
0.46 |
| chr8_13869612_13869789 | 0.15 |
Champ1 |
chromosome alignment maintaining phosphoprotein 1 |
4 |
0.96 |
| chr5_89341929_89342080 | 0.15 |
Gc |
vitamin D binding protein |
93624 |
0.09 |
| chr17_35837067_35837599 | 0.15 |
Tubb5 |
tubulin, beta 5 class I |
273 |
0.75 |
| chr19_3912063_3912250 | 0.15 |
Ndufs8 |
NADH:ubiquinone oxidoreductase core subunit S8 |
332 |
0.72 |
| chr3_84345271_84345622 | 0.15 |
4930565D16Rik |
RIKEN cDNA 4930565D16 gene |
34475 |
0.17 |
| chr2_27692549_27692919 | 0.15 |
Rxra |
retinoid X receptor alpha |
15472 |
0.24 |
| chr8_66486288_66486439 | 0.15 |
Tma16 |
translation machinery associated 16 |
134 |
0.96 |
| chr1_74723320_74723471 | 0.15 |
Cyp27a1 |
cytochrome P450, family 27, subfamily a, polypeptide 1 |
9821 |
0.12 |
| chr3_88145395_88145546 | 0.15 |
Mef2d |
myocyte enhancer factor 2D |
2896 |
0.15 |
| chr2_164785973_164786236 | 0.15 |
Snx21 |
sorting nexin family member 21 |
87 |
0.91 |
| chr12_71876146_71876310 | 0.15 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
13502 |
0.21 |
| chr5_111473745_111473896 | 0.15 |
Gm43119 |
predicted gene 43119 |
50231 |
0.12 |
| chr7_30184282_30184433 | 0.15 |
Cox7a1 |
cytochrome c oxidase subunit 7A1 |
163 |
0.86 |
| chr1_172311861_172312075 | 0.14 |
Igsf8 |
immunoglobulin superfamily, member 8 |
30 |
0.95 |
| chr13_18123081_18123244 | 0.14 |
4930448F12Rik |
RIKEN cDNA 4930448F12 gene |
22960 |
0.14 |
| chr9_66179441_66179773 | 0.14 |
Dapk2 |
death-associated protein kinase 2 |
21372 |
0.18 |
| chr17_50565884_50566049 | 0.14 |
Plcl2 |
phospholipase C-like 2 |
41028 |
0.2 |
| chr12_31655019_31655182 | 0.14 |
Cog5 |
component of oligomeric golgi complex 5 |
231 |
0.54 |
| chr12_31074834_31075017 | 0.14 |
Fam110c |
family with sequence similarity 110, member C |
1064 |
0.41 |
| chr19_6995428_6995936 | 0.14 |
Nudt22 |
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
355 |
0.48 |
| chr9_45412427_45412583 | 0.14 |
Gm48135 |
predicted gene, 48135 |
1585 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.5 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.2 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.3 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 0.2 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.2 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.9 | GO:0051875 | pigment granule localization(GO:0051875) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0051138 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.3 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0034831 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.0 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |