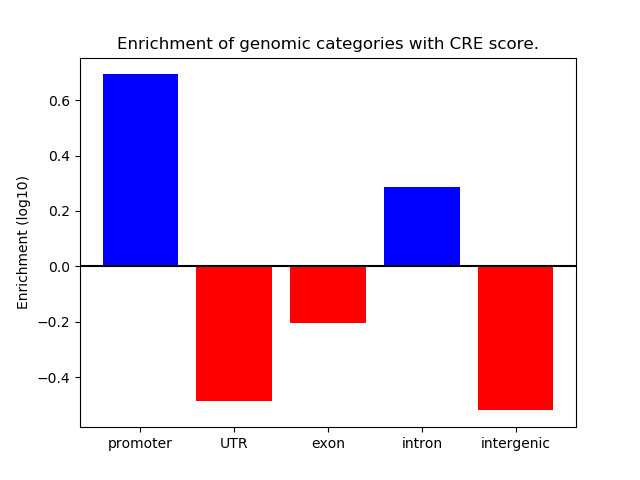
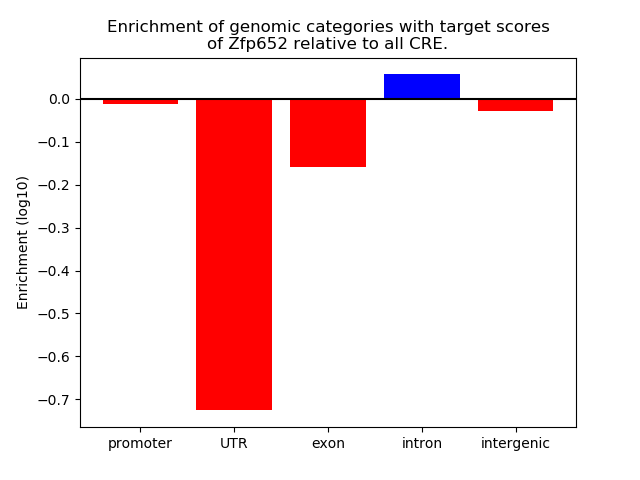
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
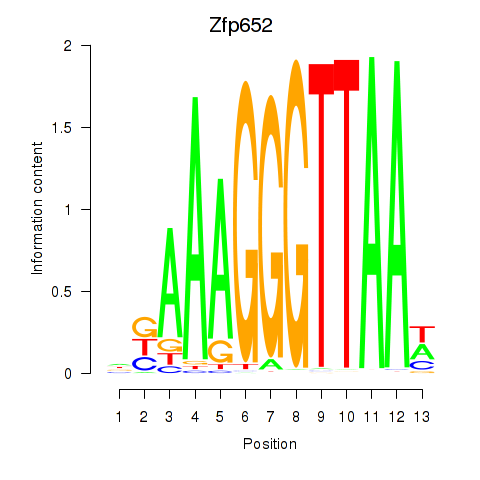
Results for Zfp652
Z-value: 2.50
Transcription factors associated with Zfp652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp652
|
ENSMUSG00000075595.3 | zinc finger protein 652 |
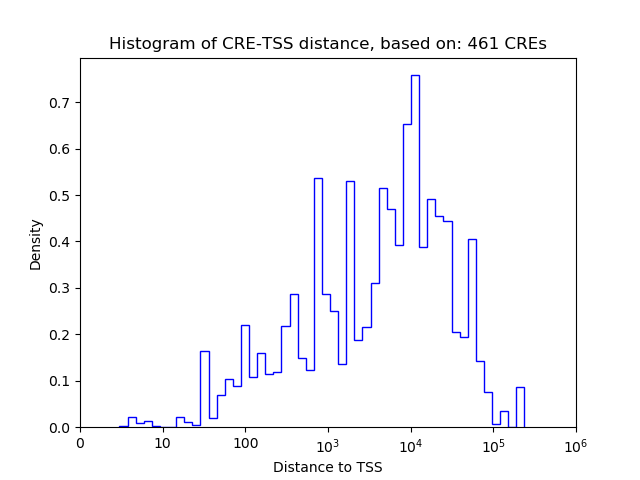
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_95709774_95709938 | Zfp652 | 2817 | 0.168699 | -0.99 | 5.4e-05 | Click! |
| chr11_95724615_95724802 | Zfp652 | 11697 | 0.122172 | -0.98 | 8.2e-04 | Click! |
| chr11_95719434_95719596 | Zfp652 | 6504 | 0.132014 | -0.97 | 1.7e-03 | Click! |
| chr11_95709547_95709712 | Zfp652 | 3044 | 0.162612 | -0.90 | 1.3e-02 | Click! |
| chr11_95750632_95750819 | Zfp652 | 1658 | 0.272858 | 0.86 | 3.0e-02 | Click! |
Activity of the Zfp652 motif across conditions
Conditions sorted by the z-value of the Zfp652 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
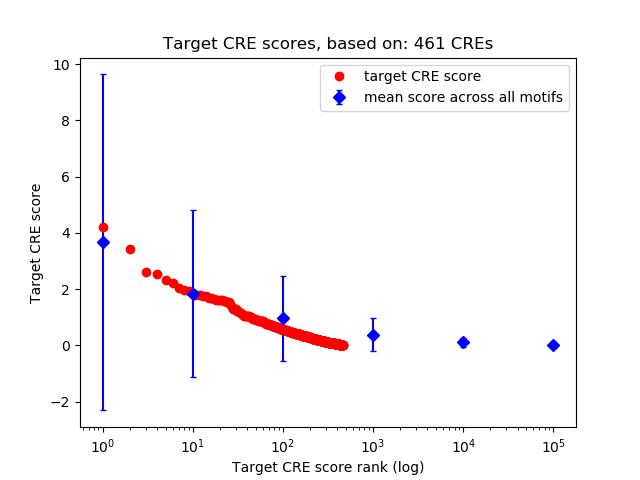
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_51751367_51751637 | 4.22 |
Gm28055 |
predicted gene 28055 |
4559 |
0.23 |
| chrX_11997001_11997158 | 3.42 |
Gm14512 |
predicted gene 14512 |
12611 |
0.24 |
| chr1_21259122_21259550 | 2.62 |
Gsta3 |
glutathione S-transferase, alpha 3 |
5815 |
0.12 |
| chr8_110644559_110644860 | 2.53 |
Vac14 |
Vac14 homolog (S. cerevisiae) |
871 |
0.63 |
| chr17_80022007_80022158 | 2.33 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr9_44087724_44088170 | 2.21 |
Usp2 |
ubiquitin specific peptidase 2 |
757 |
0.37 |
| chr1_188902459_188902627 | 2.02 |
Ush2a |
usherin |
51201 |
0.15 |
| chr14_17823512_17823678 | 1.98 |
Gm48320 |
predicted gene, 48320 |
52473 |
0.16 |
| chr1_21262098_21262745 | 1.93 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8900 |
0.11 |
| chr8_104787830_104787982 | 1.88 |
Gm45782 |
predicted gene 45782 |
2078 |
0.17 |
| chr3_89338031_89338206 | 1.78 |
Efna4 |
ephrin A4 |
90 |
0.7 |
| chr3_133726266_133726417 | 1.77 |
Gm40153 |
predicted gene, 40153 |
62950 |
0.1 |
| chr4_121078016_121078199 | 1.76 |
Zmpste24 |
zinc metallopeptidase, STE24 |
369 |
0.74 |
| chr11_77663952_77664103 | 1.74 |
Gm23132 |
predicted gene, 23132 |
5320 |
0.13 |
| chr1_21299781_21300307 | 1.69 |
Gm4956 |
predicted gene 4956 |
1732 |
0.22 |
| chr10_68126914_68127100 | 1.69 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
9619 |
0.25 |
| chr11_105353735_105353899 | 1.65 |
Gm11638 |
predicted gene 11638 |
9435 |
0.17 |
| chr9_35175169_35175403 | 1.60 |
Dcps |
decapping enzyme, scavenger |
707 |
0.52 |
| chr4_47366562_47366857 | 1.60 |
Tgfbr1 |
transforming growth factor, beta receptor I |
13099 |
0.22 |
| chr11_16826830_16827159 | 1.60 |
Egfros |
epidermal growth factor receptor, opposite strand |
3708 |
0.27 |
| chr19_26823641_26823967 | 1.60 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
103 |
0.97 |
| chr1_132476574_132476725 | 1.58 |
6030442K20Rik |
RIKEN cDNA 6030442K20 gene |
396 |
0.63 |
| chr6_94653423_94653689 | 1.56 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
1969 |
0.38 |
| chr9_68957886_68958054 | 1.54 |
Rora |
RAR-related orphan receptor alpha |
237571 |
0.02 |
| chr15_88919761_88919917 | 1.54 |
Ttll8 |
tubulin tyrosine ligase-like family, member 8 |
29 |
0.97 |
| chr19_40138748_40138899 | 1.46 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
48463 |
0.1 |
| chr6_6149940_6150263 | 1.35 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
23850 |
0.23 |
| chr2_58785851_58786309 | 1.31 |
Upp2 |
uridine phosphorylase 2 |
20755 |
0.19 |
| chr8_68082402_68082553 | 1.30 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
20191 |
0.24 |
| chr2_118532811_118533170 | 1.28 |
Bmf |
BCL2 modifying factor |
11597 |
0.16 |
| chr5_92524378_92524560 | 1.24 |
Scarb2 |
scavenger receptor class B, member 2 |
17636 |
0.15 |
| chr19_38399310_38399568 | 1.23 |
Slc35g1 |
solute carrier family 35, member G1 |
3398 |
0.2 |
| chr8_36242947_36243098 | 1.20 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
6494 |
0.21 |
| chr1_177711335_177711718 | 1.14 |
1700016C15Rik |
RIKEN cDNA 1700016C15 gene |
18288 |
0.18 |
| chr3_83048403_83048757 | 1.12 |
Fgb |
fibrinogen beta chain |
1283 |
0.37 |
| chr17_63195864_63196015 | 1.07 |
Gm24730 |
predicted gene, 24730 |
6799 |
0.29 |
| chr2_134534653_134534804 | 1.06 |
Hao1 |
hydroxyacid oxidase 1, liver |
19579 |
0.27 |
| chr14_78536823_78537132 | 1.06 |
Akap11 |
A kinase (PRKA) anchor protein 11 |
169 |
0.96 |
| chr4_82476051_82476238 | 1.05 |
Nfib |
nuclear factor I/B |
23172 |
0.18 |
| chr6_95936843_95937045 | 1.05 |
Gm7838 |
predicted gene 7838 |
10015 |
0.29 |
| chr9_105792087_105792238 | 1.05 |
Col6a6 |
collagen, type VI, alpha 6 |
17486 |
0.2 |
| chr4_106939076_106939227 | 1.03 |
Ssbp3 |
single-stranded DNA binding protein 3 |
2547 |
0.3 |
| chr6_137754656_137754862 | 1.02 |
Dera |
deoxyribose-phosphate aldolase (putative) |
144 |
0.97 |
| chr10_69584810_69585026 | 1.00 |
Gm46231 |
predicted gene, 46231 |
18557 |
0.24 |
| chr17_46026823_46026974 | 0.97 |
Vegfa |
vascular endothelial growth factor A |
2017 |
0.29 |
| chr19_7477624_7477775 | 0.97 |
Rtn3 |
reticulon 3 |
5503 |
0.15 |
| chr1_74695935_74696104 | 0.95 |
Ttll4 |
tubulin tyrosine ligase-like family, member 4 |
285 |
0.87 |
| chr18_5141716_5141913 | 0.93 |
Gm26682 |
predicted gene, 26682 |
23917 |
0.21 |
| chr4_116673334_116673488 | 0.93 |
Gm25137 |
predicted gene, 25137 |
742 |
0.5 |
| chr9_64814123_64814281 | 0.91 |
Dennd4a |
DENN/MADD domain containing 4A |
2658 |
0.3 |
| chr3_52648763_52649088 | 0.91 |
Gm10293 |
predicted pseudogene 10293 |
36090 |
0.17 |
| chr8_94871189_94871343 | 0.90 |
Dok4 |
docking protein 4 |
62 |
0.95 |
| chr3_118444081_118444268 | 0.90 |
Gm9916 |
predicted gene 9916 |
9404 |
0.15 |
| chr12_80932956_80933128 | 0.88 |
1700052I22Rik |
RIKEN cDNA 1700052I22 gene |
8593 |
0.13 |
| chr1_85105794_85106219 | 0.87 |
A530040E14Rik |
RIKEN cDNA A530040E14 gene |
60 |
0.94 |
| chr5_31494357_31494508 | 0.87 |
Gpn1 |
GPN-loop GTPase 1 |
309 |
0.68 |
| chr19_5460289_5460467 | 0.86 |
Fibp |
fibroblast growth factor (acidic) intracellular binding protein |
237 |
0.74 |
| chr4_141870823_141871025 | 0.86 |
Efhd2 |
EF hand domain containing 2 |
3996 |
0.12 |
| chr11_113140051_113140245 | 0.85 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
32929 |
0.22 |
| chr8_90947382_90947547 | 0.85 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
7494 |
0.17 |
| chr9_45918008_45918198 | 0.84 |
Pcsk7 |
proprotein convertase subtilisin/kexin type 7 |
490 |
0.62 |
| chr4_3373410_3373589 | 0.82 |
Gm11784 |
predicted gene 11784 |
13347 |
0.2 |
| chr2_94177720_94177917 | 0.80 |
Mir670hg |
MIR670 host gene (non-protein coding) |
1097 |
0.45 |
| chr19_40341299_40341486 | 0.78 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
798 |
0.65 |
| chr8_108767561_108767755 | 0.77 |
Gm38042 |
predicted gene, 38042 |
30065 |
0.2 |
| chr1_67149336_67150363 | 0.77 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
26823 |
0.2 |
| chr1_67197806_67198015 | 0.77 |
Gm15668 |
predicted gene 15668 |
51290 |
0.13 |
| chr18_38754897_38755064 | 0.76 |
Gm8302 |
predicted gene 8302 |
17367 |
0.16 |
| chr13_109485029_109485208 | 0.75 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
42935 |
0.21 |
| chr13_94806502_94806690 | 0.74 |
Tbca |
tubulin cofactor A |
11138 |
0.19 |
| chr1_36090124_36090556 | 0.74 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
11350 |
0.13 |
| chr10_7793023_7793376 | 0.74 |
Ppil4 |
peptidylprolyl isomerase (cyclophilin)-like 4 |
278 |
0.56 |
| chr1_133103203_133103354 | 0.72 |
Ppp1r15b |
protein phosphatase 1, regulatory subunit 15B |
27865 |
0.11 |
| chr4_148617730_148618042 | 0.71 |
Tardbp |
TAR DNA binding protein |
34 |
0.96 |
| chr18_64009773_64009965 | 0.71 |
Gm6974 |
predicted gene 6974 |
71551 |
0.1 |
| chr3_36533351_36533502 | 0.70 |
Gm11549 |
predicted gene 11549 |
1042 |
0.38 |
| chr8_13140413_13140571 | 0.70 |
Cul4a |
cullin 4A |
185 |
0.89 |
| chr11_98960264_98960733 | 0.69 |
Rara |
retinoic acid receptor, alpha |
86 |
0.94 |
| chr11_112055735_112055912 | 0.68 |
Gm11679 |
predicted gene 11679 |
12245 |
0.27 |
| chr13_119389612_119389948 | 0.67 |
Nnt |
nicotinamide nucleotide transhydrogenase |
19172 |
0.17 |
| chr3_52270975_52271214 | 0.67 |
Gm20402 |
predicted gene 20402 |
1659 |
0.26 |
| chr10_93094773_93094963 | 0.66 |
Gm32468 |
predicted gene, 32468 |
9617 |
0.2 |
| chr7_45127479_45128111 | 0.66 |
Rpl13a |
ribosomal protein L13A |
28 |
0.71 |
| chr15_85750165_85750444 | 0.66 |
Ppara |
peroxisome proliferator activated receptor alpha |
14173 |
0.14 |
| chr19_55107598_55107749 | 0.66 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
4582 |
0.23 |
| chr11_75049810_75049987 | 0.64 |
Gm12333 |
predicted gene 12333 |
10550 |
0.12 |
| chr4_48272049_48272240 | 0.64 |
Erp44 |
endoplasmic reticulum protein 44 |
7308 |
0.2 |
| chr1_190137352_190137664 | 0.64 |
Gm28172 |
predicted gene 28172 |
31162 |
0.16 |
| chr14_21435070_21435221 | 0.62 |
Gm25864 |
predicted gene, 25864 |
15329 |
0.19 |
| chr11_46697157_46697317 | 0.61 |
Timd2 |
T cell immunoglobulin and mucin domain containing 2 |
1725 |
0.3 |
| chr1_146494374_146494981 | 0.61 |
Brinp3 |
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
83 |
0.75 |
| chr3_102409696_102409850 | 0.61 |
Gm43245 |
predicted gene 43245 |
19766 |
0.17 |
| chr8_94966654_94966820 | 0.59 |
Gm10286 |
predicted gene 10286 |
3017 |
0.16 |
| chr1_91458255_91458771 | 0.59 |
Per2 |
period circadian clock 2 |
811 |
0.48 |
| chr7_140828126_140828283 | 0.58 |
Zfp941 |
zinc finger protein 941 |
6026 |
0.07 |
| chr18_16774605_16774780 | 0.58 |
Cdh2 |
cadherin 2 |
33770 |
0.16 |
| chr4_62699191_62699363 | 0.58 |
Rgs3 |
regulator of G-protein signaling 3 |
189 |
0.94 |
| chr14_105730837_105730988 | 0.57 |
Gm10076 |
predicted gene 10076 |
49083 |
0.13 |
| chr16_93823192_93823406 | 0.57 |
Morc3 |
microrchidia 3 |
8822 |
0.13 |
| chrX_52163913_52164117 | 0.56 |
Gpc4 |
glypican 4 |
1237 |
0.61 |
| chr2_5368187_5368380 | 0.56 |
Gm13197 |
predicted gene 13197 |
84633 |
0.09 |
| chr15_59540480_59540643 | 0.55 |
Gm36677 |
predicted gene, 36677 |
78029 |
0.09 |
| chr2_90504412_90504563 | 0.55 |
4933423P22Rik |
RIKEN cDNA 4933423P22 gene |
25237 |
0.16 |
| chr19_55743466_55744137 | 0.55 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
956 |
0.7 |
| chr3_27863709_27863878 | 0.55 |
Gm26040 |
predicted gene, 26040 |
4320 |
0.27 |
| chr15_64382862_64383209 | 0.54 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
116 |
0.97 |
| chr17_84972486_84972668 | 0.54 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
7544 |
0.18 |
| chr17_17827607_17827918 | 0.53 |
Spaca6 |
sperm acrosome associated 6 |
438 |
0.64 |
| chr17_48001818_48001969 | 0.53 |
Gm14871 |
predicted gene 14871 |
1679 |
0.3 |
| chrX_130754421_130754819 | 0.52 |
Gm26209 |
predicted gene, 26209 |
133158 |
0.05 |
| chr1_165464701_165464879 | 0.52 |
Gm37860 |
predicted gene, 37860 |
2710 |
0.13 |
| chr1_21267061_21267212 | 0.52 |
Gm28836 |
predicted gene 28836 |
4457 |
0.12 |
| chr19_25622028_25622403 | 0.51 |
Dmrt3 |
doublesex and mab-3 related transcription factor 3 |
11678 |
0.21 |
| chr2_27694949_27695100 | 0.51 |
Rxra |
retinoid X receptor alpha |
14268 |
0.25 |
| chr3_138288275_138288480 | 0.50 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
10726 |
0.12 |
| chr2_152733626_152733777 | 0.50 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
2550 |
0.16 |
| chr11_85182930_85183081 | 0.50 |
1700125H20Rik |
RIKEN cDNA 1700125H20 gene |
11909 |
0.13 |
| chr12_85304850_85305043 | 0.50 |
Zc2hc1c |
zinc finger, C2HC-type containing 1C |
16355 |
0.1 |
| chr3_94365935_94366258 | 0.49 |
C2cd4d |
C2 calcium-dependent domain containing 4D |
3593 |
0.09 |
| chr17_45594619_45594866 | 0.49 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
760 |
0.44 |
| chr17_28523067_28523337 | 0.48 |
Gm20109 |
predicted gene, 20109 |
54 |
0.53 |
| chr17_57211088_57211386 | 0.48 |
Mir6978 |
microRNA 6978 |
5991 |
0.11 |
| chr6_29828721_29828890 | 0.47 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
24955 |
0.16 |
| chr4_130721540_130721691 | 0.47 |
Snord85 |
small nucleolar RNA, C/D box 85 |
28019 |
0.12 |
| chr16_56279382_56279563 | 0.47 |
Impg2 |
interphotoreceptor matrix proteoglycan 2 |
9288 |
0.23 |
| chr8_33918433_33918601 | 0.46 |
Rbpms |
RNA binding protein gene with multiple splicing |
10759 |
0.17 |
| chr4_137795539_137795761 | 0.45 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
580 |
0.78 |
| chr14_36973905_36974284 | 0.45 |
Ccser2 |
coiled-coil serine rich 2 |
5317 |
0.22 |
| chr8_126969660_126969828 | 0.45 |
Rbm34 |
RNA binding motif protein 34 |
1305 |
0.37 |
| chr15_79321473_79321634 | 0.45 |
Pla2g6 |
phospholipase A2, group VI |
2013 |
0.19 |
| chr4_133042632_133042816 | 0.45 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
3231 |
0.25 |
| chr11_75568775_75568947 | 0.45 |
Slc43a2 |
solute carrier family 43, member 2 |
2045 |
0.17 |
| chr2_134845241_134845392 | 0.44 |
Gm14036 |
predicted gene 14036 |
41367 |
0.17 |
| chr2_158038644_158038816 | 0.43 |
Rprd1b |
regulation of nuclear pre-mRNA domain containing 1B |
4693 |
0.19 |
| chr12_54655761_54656415 | 0.43 |
Sptssa |
serine palmitoyltransferase, small subunit A |
110 |
0.91 |
| chr4_70482987_70483163 | 0.42 |
Megf9 |
multiple EGF-like-domains 9 |
51853 |
0.17 |
| chr8_111535997_111536473 | 0.42 |
Znrf1 |
zinc and ring finger 1 |
138 |
0.96 |
| chr8_41019347_41019768 | 0.42 |
B430010I23Rik |
RIKEN cDNA B430010I23 gene |
1819 |
0.27 |
| chr11_16905052_16905224 | 0.41 |
Egfr |
epidermal growth factor receptor |
47 |
0.98 |
| chr11_75297718_75298001 | 0.41 |
Gm47300 |
predicted gene, 47300 |
20749 |
0.13 |
| chr7_25280357_25280508 | 0.41 |
Cic |
capicua transcriptional repressor |
791 |
0.41 |
| chr15_81343404_81343586 | 0.41 |
Slc25a17 |
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
662 |
0.64 |
| chr4_35126863_35127042 | 0.41 |
Ifnk |
interferon kappa |
25104 |
0.16 |
| chr5_124351456_124351859 | 0.40 |
Cdk2ap1 |
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
3 |
0.96 |
| chr16_13354242_13354407 | 0.40 |
Mrtfb |
myocardin related transcription factor B |
4097 |
0.25 |
| chr10_75516024_75516215 | 0.40 |
Gucd1 |
guanylyl cyclase domain containing 1 |
1051 |
0.33 |
| chr17_26724491_26724642 | 0.39 |
Crebrf |
CREB3 regulatory factor |
8842 |
0.16 |
| chr10_23851933_23852432 | 0.39 |
Vnn3 |
vanin 3 |
720 |
0.56 |
| chr18_12075776_12075953 | 0.39 |
Tmem241 |
transmembrane protein 241 |
1233 |
0.51 |
| chr3_148680566_148680729 | 0.39 |
Gm43575 |
predicted gene 43575 |
27587 |
0.24 |
| chr13_48543479_48543831 | 0.39 |
Gm37238 |
predicted gene, 37238 |
1378 |
0.22 |
| chr13_89551647_89551801 | 0.39 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
11928 |
0.23 |
| chr6_121138262_121138418 | 0.39 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
7341 |
0.15 |
| chr2_6379003_6379169 | 0.39 |
Usp6nl |
USP6 N-terminal like |
26353 |
0.17 |
| chr4_84068033_84068731 | 0.38 |
6030471H07Rik |
RIKEN cDNA 6030471H07 gene |
23503 |
0.2 |
| chr11_98729103_98729272 | 0.38 |
Med24 |
mediator complex subunit 24 |
180 |
0.89 |
| chr12_87444275_87444437 | 0.38 |
Alkbh1 |
alkB homolog 1, histone H2A dioxygenase |
339 |
0.47 |
| chr4_76435923_76436234 | 0.36 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
13902 |
0.21 |
| chr5_108660752_108661377 | 0.36 |
Dgkq |
diacylglycerol kinase, theta |
253 |
0.76 |
| chr4_104765968_104766173 | 0.36 |
C8b |
complement component 8, beta polypeptide |
247 |
0.95 |
| chr13_95859199_95859350 | 0.35 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
32483 |
0.16 |
| chr15_57980598_57980801 | 0.35 |
Fam83a |
family with sequence similarity 83, member A |
4720 |
0.19 |
| chr10_62924201_62924495 | 0.35 |
Gm25862 |
predicted gene, 25862 |
824 |
0.43 |
| chr7_44384509_44384671 | 0.35 |
Syt3 |
synaptotagmin III |
14 |
0.92 |
| chr9_44479696_44479909 | 0.35 |
Gm47204 |
predicted gene, 47204 |
43 |
0.9 |
| chr15_83202527_83202771 | 0.35 |
7530414M10Rik |
RIKEN cDNA 7530414M10 gene |
26204 |
0.1 |
| chr19_40299087_40299297 | 0.35 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
634 |
0.57 |
| chr14_16573464_16573628 | 0.34 |
Rarb |
retinoic acid receptor, beta |
1499 |
0.4 |
| chr10_20723259_20723681 | 0.34 |
Pde7b |
phosphodiesterase 7B |
1226 |
0.53 |
| chr5_146239026_146239445 | 0.34 |
Cdk8 |
cyclin-dependent kinase 8 |
7850 |
0.12 |
| chr17_26589554_26589705 | 0.34 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
5247 |
0.13 |
| chr13_38527580_38527738 | 0.34 |
Txndc5 |
thioredoxin domain containing 5 |
255 |
0.47 |
| chr19_18146304_18146531 | 0.34 |
Gm18610 |
predicted gene, 18610 |
59378 |
0.14 |
| chr4_102811326_102811477 | 0.34 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
50876 |
0.15 |
| chr18_74764829_74765043 | 0.33 |
Scarna17 |
small Cajal body-specific RNA 17 |
13533 |
0.14 |
| chr12_101766870_101767053 | 0.33 |
Tc2n |
tandem C2 domains, nuclear |
48438 |
0.12 |
| chr2_116983226_116983382 | 0.33 |
Gm29340 |
predicted gene 29340 |
6876 |
0.2 |
| chr7_127614456_127614767 | 0.33 |
Zfp629 |
zinc finger protein 629 |
178 |
0.86 |
| chr2_167348215_167348655 | 0.33 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
748 |
0.67 |
| chr16_36935496_36935647 | 0.32 |
Hcls1 |
hematopoietic cell specific Lyn substrate 1 |
588 |
0.62 |
| chr9_119308875_119309095 | 0.32 |
Gm22729 |
predicted gene, 22729 |
2228 |
0.21 |
| chr11_98796894_98797678 | 0.32 |
Msl1 |
male specific lethal 1 |
1368 |
0.26 |
| chr11_77661061_77661234 | 0.32 |
Gm23132 |
predicted gene, 23132 |
2440 |
0.19 |
| chr6_124541506_124541699 | 0.32 |
C1s1 |
complement component 1, s subcomponent 1 |
701 |
0.5 |
| chr6_137738154_137738336 | 0.31 |
Strap |
serine/threonine kinase receptor associated protein |
3167 |
0.29 |
| chr10_9009141_9009301 | 0.30 |
Gm48728 |
predicted gene, 48728 |
57272 |
0.12 |
| chr11_34181831_34182010 | 0.30 |
Foxi1 |
forkhead box I1 |
26169 |
0.16 |
| chr13_48540722_48541023 | 0.30 |
Mirlet7a-1 |
microRNA let7a-1 |
2600 |
0.12 |
| chr8_117737291_117737442 | 0.30 |
Gm31774 |
predicted gene, 31774 |
10894 |
0.14 |
| chr7_119847929_119848089 | 0.29 |
Rexo5 |
RNA exonuclease 5 |
3803 |
0.17 |
| chr11_119577189_119577358 | 0.29 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
25632 |
0.13 |
| chr18_80294645_80295296 | 0.29 |
Gm37015 |
predicted gene, 37015 |
8710 |
0.13 |
| chr6_36175492_36175671 | 0.29 |
Gm43443 |
predicted gene 43443 |
13518 |
0.28 |
| chr7_3073209_3073392 | 0.29 |
Gm3104 |
predicted gene 3104 |
12701 |
0.1 |
| chr1_151132224_151132407 | 0.29 |
Gm8941 |
predicted gene 8941 |
4192 |
0.14 |
| chr4_82447093_82447266 | 0.29 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
7769 |
0.24 |
| chr1_64533839_64534218 | 0.28 |
Creb1 |
cAMP responsive element binding protein 1 |
1134 |
0.54 |
| chr6_72117206_72117587 | 0.28 |
St3gal5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
2164 |
0.2 |
| chr5_64506002_64506172 | 0.28 |
C030018K13Rik |
RIKEN cDNA C030018K13 gene |
29052 |
0.1 |
| chr4_152229764_152230378 | 0.28 |
Gm13096 |
predicted gene 13096 |
26145 |
0.1 |
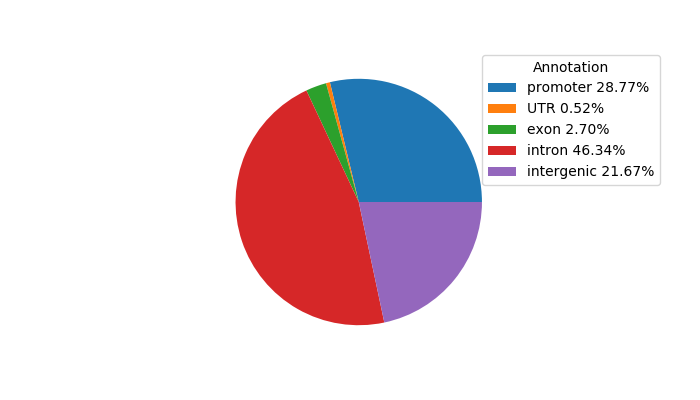
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.4 | 1.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.3 | 1.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.2 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.5 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.7 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 0.3 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.1 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.4 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.2 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.1 | 0.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 1.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.3 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.5 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0036492 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) regulation of translation initiation in response to endoplasmic reticulum stress(GO:0036491) eiF2alpha phosphorylation in response to endoplasmic reticulum stress(GO:0036492) |
| 0.0 | 0.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.0 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.2 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.0 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.2 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.0 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.0 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.1 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 1.5 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.0 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 1.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 1.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 1.5 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |