Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
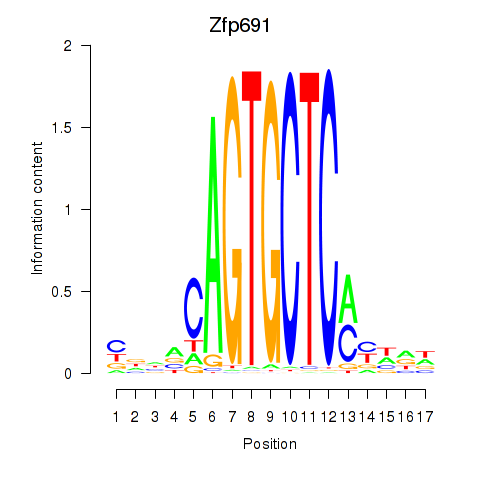
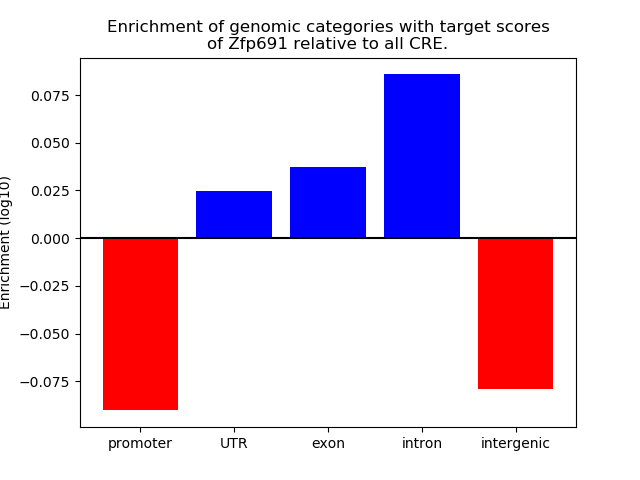
Results for Zfp691
Z-value: 1.03
Transcription factors associated with Zfp691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp691
|
ENSMUSG00000045268.7 | zinc finger protein 691 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_119174225_119174388 | Zfp691 | 111 | 0.925119 | -0.62 | 1.9e-01 | Click! |
| chr4_119167778_119167932 | Zfp691 | 6001 | 0.097115 | 0.53 | 2.8e-01 | Click! |
| chr4_119174501_119174685 | Zfp691 | 398 | 0.710384 | 0.41 | 4.2e-01 | Click! |
| chr4_119173732_119174046 | Zfp691 | 33 | 0.947957 | 0.23 | 6.7e-01 | Click! |
Activity of the Zfp691 motif across conditions
Conditions sorted by the z-value of the Zfp691 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
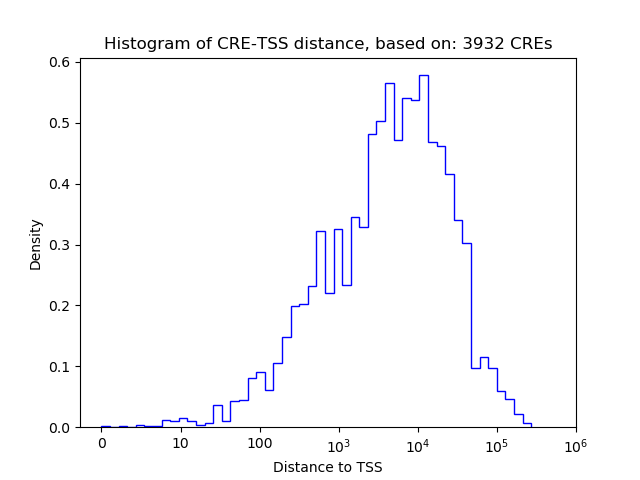
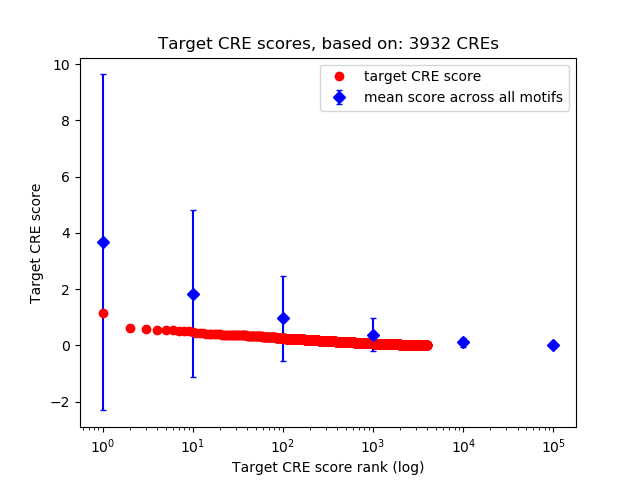
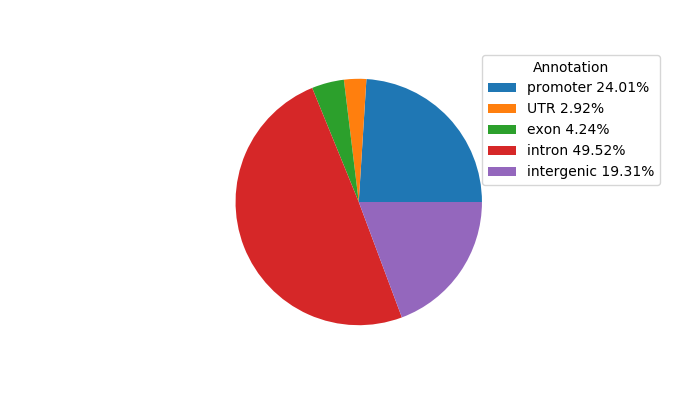
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_97413837_97414007 | 1.13 |
Thrsp |
thyroid hormone responsive |
3597 |
0.16 |
| chr3_51255928_51256079 | 0.62 |
Elf2 |
E74-like factor 2 |
4238 |
0.15 |
| chr4_88912975_88913126 | 0.58 |
Mir31 |
microRNA 31 |
2388 |
0.15 |
| chr10_89516308_89516459 | 0.56 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
9725 |
0.22 |
| chr3_149437041_149437628 | 0.53 |
Gm30382 |
predicted gene, 30382 |
7942 |
0.29 |
| chr1_100298140_100298304 | 0.53 |
Gm29667 |
predicted gene 29667 |
15431 |
0.18 |
| chr12_104346326_104346873 | 0.52 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
8113 |
0.12 |
| chr19_44403590_44403745 | 0.52 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
3023 |
0.2 |
| chr8_46902999_46903150 | 0.49 |
1700011L03Rik |
RIKEN cDNA 1700011L03 gene |
45725 |
0.12 |
| chr2_33447436_33447587 | 0.46 |
Gm13536 |
predicted gene 13536 |
709 |
0.62 |
| chr6_143243938_143244283 | 0.43 |
D6Ertd474e |
DNA segment, Chr 6, ERATO Doi 474, expressed |
1778 |
0.37 |
| chr4_6268102_6268325 | 0.42 |
Gm11798 |
predicted gene 11798 |
7248 |
0.2 |
| chr11_16822013_16822228 | 0.42 |
Egfros |
epidermal growth factor receptor, opposite strand |
8582 |
0.23 |
| chr8_122047070_122047266 | 0.42 |
Banp |
BTG3 associated nuclear protein |
46321 |
0.11 |
| chr11_115165779_115165997 | 0.42 |
Slc9a3r1 |
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
2547 |
0.17 |
| chr17_44943829_44944027 | 0.41 |
4930564C03Rik |
RIKEN cDNA 4930564C03 gene |
63633 |
0.13 |
| chr18_9503676_9503906 | 0.40 |
Gm7527 |
predicted gene 7527 |
8170 |
0.15 |
| chr3_18222967_18223118 | 0.40 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
20296 |
0.2 |
| chr7_112449712_112449880 | 0.39 |
Parva |
parvin, alpha |
21891 |
0.21 |
| chr11_120813632_120813783 | 0.39 |
Fasn |
fatty acid synthase |
1748 |
0.19 |
| chr18_75504426_75504613 | 0.38 |
Gm10532 |
predicted gene 10532 |
10126 |
0.27 |
| chr18_75497746_75497943 | 0.38 |
Gm10532 |
predicted gene 10532 |
16801 |
0.25 |
| chr19_55394273_55394505 | 0.38 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
13375 |
0.25 |
| chr4_140950673_140950824 | 0.38 |
Padi2 |
peptidyl arginine deiminase, type II |
4336 |
0.13 |
| chr9_42464224_42464407 | 0.38 |
Tbcel |
tubulin folding cofactor E-like |
2854 |
0.24 |
| chr8_79710908_79711237 | 0.38 |
Abce1 |
ATP-binding cassette, sub-family E (OABP), member 1 |
578 |
0.48 |
| chr16_93860540_93860703 | 0.37 |
Morc3 |
microrchidia 3 |
108 |
0.95 |
| chr17_71214957_71215123 | 0.37 |
Lpin2 |
lipin 2 |
10364 |
0.17 |
| chr10_44765375_44765553 | 0.37 |
4930431F10Rik |
RIKEN cDNA 4930431F10 gene |
26564 |
0.12 |
| chr15_55291670_55291894 | 0.37 |
Col14a1 |
collagen, type XIV, alpha 1 |
15968 |
0.21 |
| chr19_58369154_58369339 | 0.36 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
85220 |
0.09 |
| chr9_107316197_107316355 | 0.36 |
Hemk1 |
HemK methyltransferase family member 1 |
13176 |
0.09 |
| chr17_31036229_31036438 | 0.36 |
Abcg1 |
ATP binding cassette subfamily G member 1 |
21342 |
0.11 |
| chr17_32944138_32944328 | 0.36 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
3102 |
0.14 |
| chr3_89133157_89133825 | 0.36 |
Pklr |
pyruvate kinase liver and red blood cell |
2651 |
0.11 |
| chr3_118606217_118606485 | 0.36 |
Dpyd |
dihydropyrimidine dehydrogenase |
44165 |
0.15 |
| chr11_5706948_5707147 | 0.36 |
Mrps24 |
mitochondrial ribosomal protein S24 |
595 |
0.67 |
| chr5_33462368_33462568 | 0.35 |
Gm43851 |
predicted gene 43851 |
25004 |
0.15 |
| chr15_85766228_85766381 | 0.35 |
Ppara |
peroxisome proliferator activated receptor alpha |
5910 |
0.16 |
| chr15_7145941_7146124 | 0.35 |
Lifr |
LIF receptor alpha |
5483 |
0.3 |
| chr9_119151307_119151477 | 0.35 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
1329 |
0.3 |
| chr11_118508000_118508168 | 0.35 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
731 |
0.62 |
| chr16_3236787_3236938 | 0.35 |
Gm23215 |
predicted gene, 23215 |
12722 |
0.18 |
| chr2_18432844_18433010 | 0.34 |
Dnajc1 |
DnaJ heat shock protein family (Hsp40) member C1 |
40097 |
0.17 |
| chr17_79881115_79881410 | 0.34 |
Atl2 |
atlastin GTPase 2 |
14789 |
0.16 |
| chr2_4565728_4565886 | 0.34 |
Frmd4a |
FERM domain containing 4A |
369 |
0.88 |
| chr16_26643591_26643759 | 0.33 |
Il1rap |
interleukin 1 receptor accessory protein |
19519 |
0.26 |
| chr13_12597462_12597622 | 0.33 |
Gm25483 |
predicted gene, 25483 |
7977 |
0.13 |
| chr15_88832888_88833102 | 0.33 |
Gm23144 |
predicted gene, 23144 |
2305 |
0.21 |
| chr7_99139312_99140124 | 0.33 |
Uvrag |
UV radiation resistance associated gene |
1400 |
0.28 |
| chr4_3083393_3083799 | 0.33 |
Vmn1r-ps2 |
vomeronasal 1 receptor, pseudogene 2 |
20996 |
0.18 |
| chr18_9504029_9504426 | 0.32 |
Gm7527 |
predicted gene 7527 |
8606 |
0.15 |
| chr2_72227448_72227599 | 0.32 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
4779 |
0.19 |
| chr9_15303135_15303291 | 0.32 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
1657 |
0.11 |
| chr1_86951699_86951911 | 0.32 |
Gm37967 |
predicted gene, 37967 |
33813 |
0.09 |
| chr2_90958033_90958224 | 0.32 |
Celf1 |
CUGBP, Elav-like family member 1 |
6964 |
0.13 |
| chr1_67201067_67201218 | 0.31 |
Gm15668 |
predicted gene 15668 |
48058 |
0.14 |
| chr1_91460257_91460423 | 0.31 |
Per2 |
period circadian clock 2 |
1016 |
0.39 |
| chr2_48403972_48404299 | 0.31 |
Gm13472 |
predicted gene 13472 |
16098 |
0.2 |
| chr4_99033022_99033203 | 0.31 |
Angptl3 |
angiopoietin-like 3 |
67 |
0.97 |
| chr6_51678830_51679023 | 0.31 |
Gm38811 |
predicted gene, 38811 |
32155 |
0.18 |
| chr4_139629117_139629268 | 0.31 |
Aldh4a1 |
aldehyde dehydrogenase 4 family, member A1 |
6083 |
0.15 |
| chr6_117905744_117905933 | 0.31 |
Hnrnpf |
heterogeneous nuclear ribonucleoprotein F |
952 |
0.37 |
| chr4_53475197_53475646 | 0.31 |
Slc44a1 |
solute carrier family 44, member 1 |
34737 |
0.17 |
| chr3_97639633_97639825 | 0.31 |
Fmo5 |
flavin containing monooxygenase 5 |
10846 |
0.13 |
| chr11_120809069_120809447 | 0.30 |
Fasn |
fatty acid synthase |
443 |
0.66 |
| chr19_58356888_58357039 | 0.30 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
97503 |
0.07 |
| chr1_162963987_162964158 | 0.30 |
Gm37273 |
predicted gene, 37273 |
18942 |
0.14 |
| chr15_55046996_55047237 | 0.29 |
Taf2 |
TATA-box binding protein associated factor 2 |
950 |
0.51 |
| chr9_75444782_75444933 | 0.29 |
Leo1 |
Leo1, Paf1/RNA polymerase II complex component |
3321 |
0.16 |
| chr11_40754622_40755130 | 0.29 |
Ccng1 |
cyclin G1 |
435 |
0.82 |
| chr12_80824527_80824832 | 0.29 |
Susd6 |
sushi domain containing 6 |
34120 |
0.11 |
| chr11_120467825_120468138 | 0.29 |
Hgs |
HGF-regulated tyrosine kinase substrate |
346 |
0.43 |
| chr19_58349019_58349170 | 0.29 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
105372 |
0.07 |
| chr10_121457902_121458053 | 0.29 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
4236 |
0.14 |
| chr6_149168171_149168324 | 0.29 |
Amn1 |
antagonist of mitotic exit network 1 |
5124 |
0.16 |
| chr4_122983797_122983948 | 0.29 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
11780 |
0.13 |
| chr11_118739241_118739392 | 0.28 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
21725 |
0.2 |
| chr14_121720285_121720661 | 0.28 |
Dock9 |
dedicator of cytokinesis 9 |
18388 |
0.23 |
| chr9_113793219_113793637 | 0.28 |
Clasp2 |
CLIP associating protein 2 |
93 |
0.97 |
| chr12_28038561_28038854 | 0.28 |
Gm25923 |
predicted gene, 25923 |
21821 |
0.21 |
| chr5_39163040_39163219 | 0.28 |
Gm40293 |
predicted gene, 40293 |
143955 |
0.04 |
| chr8_121980914_121981086 | 0.28 |
Banp |
BTG3 associated nuclear protein |
2678 |
0.16 |
| chr10_81412487_81412833 | 0.27 |
Gm16104 |
predicted gene 16104 |
3085 |
0.08 |
| chr1_23269941_23270092 | 0.27 |
Mir30a |
microRNA 30a |
2253 |
0.2 |
| chr14_18375685_18375951 | 0.27 |
Ube2e1 |
ubiquitin-conjugating enzyme E2E 1 |
43959 |
0.14 |
| chr10_77441540_77441756 | 0.27 |
Gm35920 |
predicted gene, 35920 |
16269 |
0.15 |
| chr19_34095441_34095798 | 0.27 |
Lipm |
lipase, family member M |
5314 |
0.16 |
| chr9_70835523_70835734 | 0.26 |
Gm3436 |
predicted pseudogene 3436 |
16948 |
0.19 |
| chr2_58756221_58756376 | 0.26 |
Upp2 |
uridine phosphorylase 2 |
1086 |
0.54 |
| chr8_25776385_25776540 | 0.26 |
Bag4 |
BCL2-associated athanogene 4 |
1120 |
0.32 |
| chr3_145924039_145924997 | 0.26 |
Bcl10 |
B cell leukemia/lymphoma 10 |
161 |
0.95 |
| chr1_87572169_87572320 | 0.26 |
Ngef |
neuronal guanine nucleotide exchange factor |
1626 |
0.26 |
| chr10_80667433_80667584 | 0.25 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
4468 |
0.1 |
| chr9_74878263_74878414 | 0.25 |
Onecut1 |
one cut domain, family member 1 |
11854 |
0.15 |
| chr9_111441745_111441954 | 0.25 |
Dclk3 |
doublecortin-like kinase 3 |
2768 |
0.28 |
| chr11_16829553_16829792 | 0.25 |
Egfros |
epidermal growth factor receptor, opposite strand |
1030 |
0.59 |
| chr9_48984872_48985421 | 0.25 |
Usp28 |
ubiquitin specific peptidase 28 |
229 |
0.92 |
| chr15_96930772_96930923 | 0.25 |
Rpl10a-ps3 |
ribosomal protein L10A, pseudogene 3 |
7442 |
0.28 |
| chr13_108289333_108289486 | 0.25 |
Gm8990 |
predicted gene 8990 |
21341 |
0.14 |
| chr3_51253474_51253774 | 0.25 |
Elf2 |
E74-like factor 2 |
6617 |
0.13 |
| chr12_30334375_30334538 | 0.24 |
Sntg2 |
syntrophin, gamma 2 |
23227 |
0.23 |
| chr1_160280517_160280905 | 0.24 |
Rabgap1l |
RAB GTPase activating protein 1-like |
6627 |
0.14 |
| chr12_72476829_72477076 | 0.24 |
Lrrc9 |
leucine rich repeat containing 9 |
878 |
0.43 |
| chr7_141304868_141305072 | 0.24 |
Deaf1 |
DEAF1, transcription factor |
5950 |
0.08 |
| chr15_54587350_54587502 | 0.24 |
Mal2 |
mal, T cell differentiation protein 2 |
16234 |
0.24 |
| chrX_60400421_60400586 | 0.24 |
Atp11c |
ATPase, class VI, type 11C |
3478 |
0.24 |
| chr9_16216215_16216414 | 0.24 |
Fat3 |
FAT atypical cadherin 3 |
161917 |
0.04 |
| chr7_49525838_49525989 | 0.24 |
Nav2 |
neuron navigator 2 |
22279 |
0.22 |
| chr13_63562151_63562325 | 0.24 |
Ptch1 |
patched 1 |
1577 |
0.3 |
| chr11_108378006_108378158 | 0.24 |
Gm11656 |
predicted gene 11656 |
189 |
0.93 |
| chr1_74718194_74718570 | 0.24 |
Cyp27a1 |
cytochrome P450, family 27, subfamily a, polypeptide 1 |
4808 |
0.14 |
| chr15_89480604_89481054 | 0.24 |
C230037L18Rik |
RIKEN cDNA C230037L18 gene |
3090 |
0.12 |
| chr4_81673780_81673993 | 0.24 |
Gm11411 |
predicted gene 11411 |
22555 |
0.24 |
| chr17_86283565_86284088 | 0.24 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
3352 |
0.34 |
| chr4_83244037_83244213 | 0.24 |
Ttc39b |
tetratricopeptide repeat domain 39B |
10955 |
0.2 |
| chr15_38057614_38058038 | 0.23 |
Gm22353 |
predicted gene, 22353 |
459 |
0.79 |
| chr9_44078939_44079310 | 0.23 |
Usp2 |
ubiquitin specific peptidase 2 |
5815 |
0.08 |
| chr13_112123093_112123287 | 0.23 |
Gm31104 |
predicted gene, 31104 |
14926 |
0.2 |
| chr6_134473644_134473820 | 0.23 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
2439 |
0.29 |
| chr10_89467439_89467590 | 0.23 |
Gas2l3 |
growth arrest-specific 2 like 3 |
23547 |
0.19 |
| chr15_3574518_3574701 | 0.23 |
Ghr |
growth hormone receptor |
7233 |
0.26 |
| chr9_46111267_46111456 | 0.23 |
Sik3 |
SIK family kinase 3 |
11769 |
0.18 |
| chr6_84544470_84544631 | 0.23 |
4930504D19Rik |
RIKEN cDNA 4930504D19 gene |
8674 |
0.25 |
| chr5_100379090_100379436 | 0.23 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
264 |
0.9 |
| chr19_59343334_59343485 | 0.23 |
Pdzd8 |
PDZ domain containing 8 |
2371 |
0.23 |
| chr18_62069608_62069769 | 0.23 |
Gm41750 |
predicted gene, 41750 |
72930 |
0.09 |
| chr2_177822071_177822226 | 0.23 |
Gm14325 |
predicted gene 14325 |
12498 |
0.17 |
| chr9_65427730_65427881 | 0.23 |
Slc51b |
solute carrier family 51, beta subunit |
4850 |
0.11 |
| chr4_45341701_45341886 | 0.23 |
Dcaf10 |
DDB1 and CUL4 associated factor 10 |
308 |
0.76 |
| chr17_84990583_84990745 | 0.23 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
3009 |
0.24 |
| chr18_9503170_9503342 | 0.23 |
Gm7527 |
predicted gene 7527 |
7635 |
0.15 |
| chr19_10098484_10098639 | 0.23 |
Fads2 |
fatty acid desaturase 2 |
3185 |
0.19 |
| chr5_124234244_124234932 | 0.23 |
Gm42425 |
predicted gene 42425 |
3136 |
0.14 |
| chr10_42189906_42190057 | 0.23 |
Foxo3 |
forkhead box O3 |
68385 |
0.11 |
| chr12_111759014_111759441 | 0.23 |
Klc1 |
kinesin light chain 1 |
214 |
0.89 |
| chr4_59527591_59527772 | 0.23 |
Gm12529 |
predicted gene 12529 |
5066 |
0.18 |
| chr10_69219390_69219541 | 0.22 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
80 |
0.97 |
| chr15_25692632_25692796 | 0.22 |
Gm2862 |
predicted gene 2862 |
10916 |
0.2 |
| chr9_122164867_122165083 | 0.22 |
Snrk |
SNF related kinase |
1106 |
0.4 |
| chr13_45827730_45827889 | 0.22 |
Atxn1 |
ataxin 1 |
44479 |
0.17 |
| chr4_150433872_150434155 | 0.22 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
27619 |
0.2 |
| chr4_120668847_120669023 | 0.22 |
Cited4 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
2363 |
0.24 |
| chr11_52364094_52364245 | 0.22 |
Vdac1 |
voltage-dependent anion channel 1 |
3034 |
0.23 |
| chr6_121302765_121302947 | 0.22 |
Slc6a13 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
2074 |
0.24 |
| chr19_59343091_59343283 | 0.22 |
Pdzd8 |
PDZ domain containing 8 |
2593 |
0.21 |
| chr5_135168116_135168607 | 0.22 |
Bcl7b |
B cell CLL/lymphoma 7B |
28 |
0.96 |
| chr7_101344939_101345805 | 0.22 |
Gm45620 |
predicted gene 45620 |
2612 |
0.13 |
| chr1_87572332_87572542 | 0.22 |
Ngef |
neuronal guanine nucleotide exchange factor |
1433 |
0.28 |
| chr1_184054223_184054474 | 0.22 |
Dusp10 |
dual specificity phosphatase 10 |
19967 |
0.21 |
| chr11_115699012_115699623 | 0.22 |
Grb2 |
growth factor receptor bound protein 2 |
10 |
0.96 |
| chr3_146501639_146502276 | 0.22 |
Gng5 |
guanine nucleotide binding protein (G protein), gamma 5 |
1848 |
0.22 |
| chr5_107909531_107909737 | 0.22 |
Gm26387 |
predicted gene, 26387 |
2289 |
0.16 |
| chr14_21304994_21305440 | 0.22 |
Adk |
adenosine kinase |
12908 |
0.27 |
| chr10_17562788_17562978 | 0.21 |
Gm47770 |
predicted gene, 47770 |
35969 |
0.15 |
| chr3_98644806_98645013 | 0.21 |
Hsd3b5 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
14657 |
0.13 |
| chr15_41831558_41831716 | 0.21 |
Oxr1 |
oxidation resistance 1 |
644 |
0.74 |
| chr12_71876146_71876310 | 0.21 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
13502 |
0.21 |
| chr1_36088562_36088713 | 0.21 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
13053 |
0.13 |
| chr4_132042022_132042208 | 0.21 |
Gm13063 |
predicted gene 13063 |
6063 |
0.13 |
| chr6_94083257_94083610 | 0.21 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
169743 |
0.03 |
| chr11_16826448_16826767 | 0.21 |
Egfros |
epidermal growth factor receptor, opposite strand |
4095 |
0.26 |
| chr10_120306126_120306277 | 0.21 |
Gm38141 |
predicted gene, 38141 |
4077 |
0.18 |
| chr8_34804942_34805163 | 0.21 |
Dusp4 |
dual specificity phosphatase 4 |
2245 |
0.35 |
| chr12_81892075_81892243 | 0.21 |
Pcnx |
pecanex homolog |
26484 |
0.19 |
| chr18_9492931_9493238 | 0.21 |
Gm7527 |
predicted gene 7527 |
2537 |
0.23 |
| chr19_59942843_59943012 | 0.21 |
Rab11fip2 |
RAB11 family interacting protein 2 (class I) |
73 |
0.94 |
| chr4_53204327_53204492 | 0.21 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
13152 |
0.18 |
| chr5_114155293_114155464 | 0.21 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
8843 |
0.12 |
| chr8_46747891_46748042 | 0.21 |
Irf2 |
interferon regulatory factor 2 |
7700 |
0.15 |
| chr19_8400709_8400872 | 0.21 |
Slc22a30 |
solute carrier family 22, member 30 |
4270 |
0.24 |
| chr19_29520625_29520778 | 0.21 |
A930007I19Rik |
RIKEN cDNA A930007I19 gene |
269 |
0.89 |
| chr3_97653476_97653627 | 0.21 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
4642 |
0.15 |
| chr4_41144518_41144669 | 0.21 |
Ube2r2 |
ubiquitin-conjugating enzyme E2R 2 |
8850 |
0.1 |
| chr7_81002031_81002182 | 0.21 |
Gm44967 |
predicted gene 44967 |
7423 |
0.11 |
| chr4_108055876_108056241 | 0.20 |
Scp2 |
sterol carrier protein 2, liver |
8179 |
0.14 |
| chr16_59403797_59403956 | 0.20 |
Gabrr3 |
gamma-aminobutyric acid (GABA) receptor, rho 3 |
3456 |
0.19 |
| chr9_121354050_121354226 | 0.20 |
Gm47092 |
predicted gene, 47092 |
7582 |
0.17 |
| chr5_28075036_28075588 | 0.20 |
Insig1 |
insulin induced gene 1 |
646 |
0.68 |
| chr17_14960075_14960238 | 0.20 |
Phf10 |
PHD finger protein 10 |
629 |
0.58 |
| chr18_59108846_59109012 | 0.20 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
46418 |
0.17 |
| chr14_25492100_25492326 | 0.20 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
10442 |
0.14 |
| chr4_73044651_73044846 | 0.20 |
Gm11251 |
predicted gene 11251 |
14929 |
0.25 |
| chr5_24387652_24387803 | 0.20 |
Atg9b |
autophagy related 9B |
239 |
0.82 |
| chr5_77314693_77314994 | 0.20 |
Gm42758 |
predicted gene 42758 |
3812 |
0.15 |
| chr9_57748270_57748461 | 0.20 |
Clk3 |
CDC-like kinase 3 |
5096 |
0.15 |
| chr19_46197580_46197759 | 0.20 |
Gm8974 |
predicted gene 8974 |
8189 |
0.14 |
| chr15_85678642_85678813 | 0.20 |
Lncppara |
long noncoding RNA near Ppara |
25046 |
0.12 |
| chr16_32608318_32608521 | 0.20 |
Tfrc |
transferrin receptor |
501 |
0.74 |
| chr18_39241272_39241487 | 0.20 |
Arhgap26 |
Rho GTPase activating protein 26 |
14011 |
0.24 |
| chr16_46623333_46623484 | 0.20 |
Gm17900 |
predicted gene, 17900 |
85573 |
0.1 |
| chr8_36715111_36715588 | 0.20 |
Dlc1 |
deleted in liver cancer 1 |
17705 |
0.27 |
| chr8_67923947_67924108 | 0.19 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
13095 |
0.23 |
| chr4_120406128_120406641 | 0.19 |
Scmh1 |
sex comb on midleg homolog 1 |
956 |
0.65 |
| chr15_39907114_39907454 | 0.19 |
Lrp12 |
low density lipoprotein-related protein 12 |
36406 |
0.14 |
| chr4_152228324_152228477 | 0.19 |
Gm13096 |
predicted gene 13096 |
24474 |
0.11 |
| chr14_34328663_34328894 | 0.19 |
Glud1 |
glutamate dehydrogenase 1 |
116 |
0.93 |
| chr11_120814981_120815132 | 0.19 |
Fasn |
fatty acid synthase |
399 |
0.71 |
| chr17_26512736_26512906 | 0.19 |
Dusp1 |
dual specificity phosphatase 1 |
4302 |
0.12 |
| chr13_13337377_13337528 | 0.19 |
Gm25804 |
predicted gene, 25804 |
7968 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.2 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.1 | 0.2 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.0 | GO:2000981 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:2000489 | regulation of hepatic stellate cell activation(GO:2000489) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.0 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0000701 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |