Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
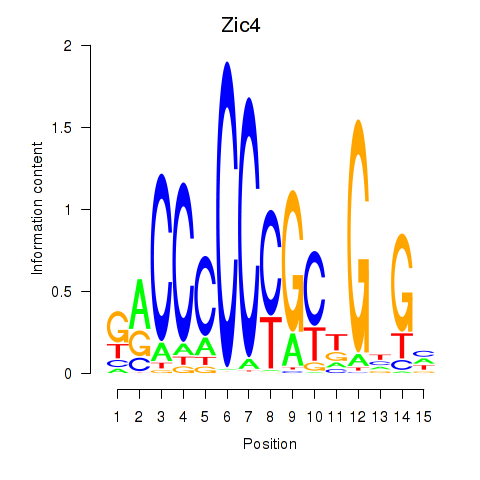
Results for Zic4
Z-value: 1.81
Transcription factors associated with Zic4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic4
|
ENSMUSG00000036972.8 | zinc finger protein of the cerebellum 4 |
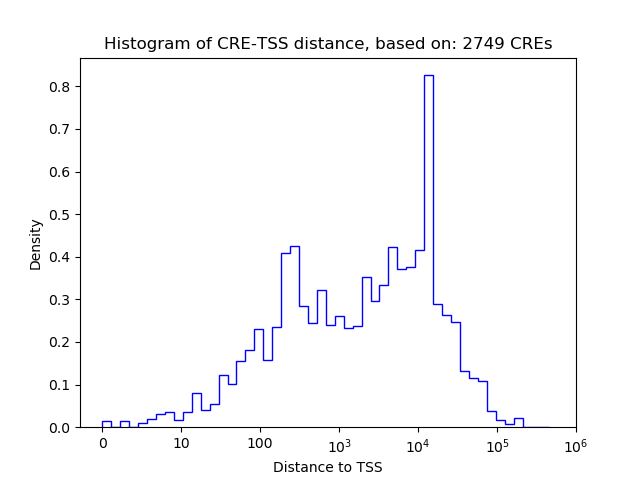
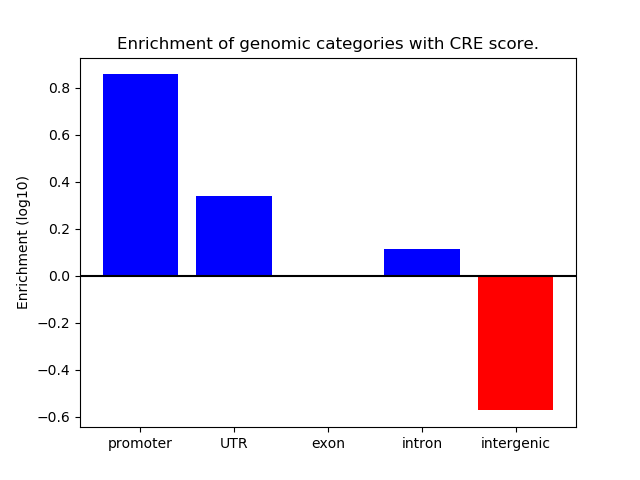
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_91370473_91370649 | Zic4 | 443 | 0.716223 | -0.27 | 6.0e-01 | Click! |
| chr9_91362060_91362437 | Zic4 | 165 | 0.856198 | 0.22 | 6.7e-01 | Click! |
Activity of the Zic4 motif across conditions
Conditions sorted by the z-value of the Zic4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
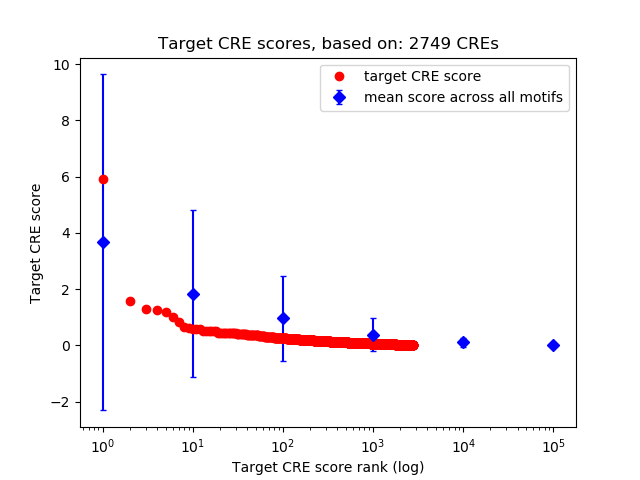
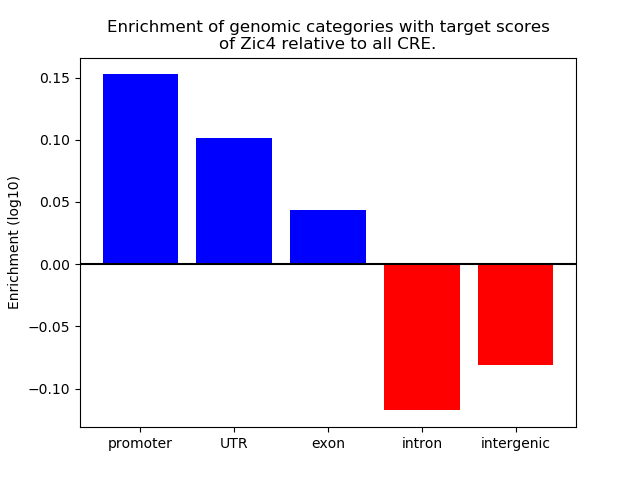
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_60681580_60681975 | 5.93 |
Dapk1 |
death associated protein kinase 1 |
14636 |
0.17 |
| chr13_60680911_60681084 | 1.59 |
Dapk1 |
death associated protein kinase 1 |
15416 |
0.17 |
| chr13_60681310_60681461 | 1.29 |
Dapk1 |
death associated protein kinase 1 |
15028 |
0.17 |
| chr6_72607199_72607442 | 1.25 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
675 |
0.43 |
| chr13_10355578_10355729 | 1.18 |
Gm26861 |
predicted gene, 26861 |
1942 |
0.37 |
| chr7_13007324_13007475 | 1.01 |
Zbtb45 |
zinc finger and BTB domain containing 45 |
2401 |
0.12 |
| chr13_60682029_60682204 | 0.83 |
Dapk1 |
death associated protein kinase 1 |
14297 |
0.17 |
| chr12_24651113_24651273 | 0.64 |
Klf11 |
Kruppel-like factor 11 |
81 |
0.96 |
| chr6_90736325_90736476 | 0.63 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
287 |
0.9 |
| chr7_127804168_127804541 | 0.59 |
9430064I24Rik |
RIKEN cDNA 9430064I24 gene |
1592 |
0.16 |
| chr11_102777578_102777754 | 0.57 |
Adam11 |
a disintegrin and metallopeptidase domain 11 |
1112 |
0.37 |
| chr13_54081500_54081675 | 0.57 |
Sfxn1 |
sideroflexin 1 |
4359 |
0.22 |
| chr12_84221379_84221549 | 0.52 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
2583 |
0.16 |
| chr4_144959729_144959958 | 0.51 |
Gm38074 |
predicted gene, 38074 |
995 |
0.56 |
| chr4_149980056_149980645 | 0.51 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
20915 |
0.13 |
| chr7_3331834_3332020 | 0.50 |
Cacng7 |
calcium channel, voltage-dependent, gamma subunit 7 |
1028 |
0.31 |
| chr1_133527738_133527935 | 0.50 |
Gm8596 |
predicted gene 8596 |
1643 |
0.39 |
| chr15_67420672_67420895 | 0.49 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
194014 |
0.03 |
| chr18_12644066_12644542 | 0.45 |
Gm50063 |
predicted gene, 50063 |
60 |
0.94 |
| chr11_51915955_51916321 | 0.45 |
Gm39822 |
predicted gene, 39822 |
17172 |
0.14 |
| chr19_6401505_6401681 | 0.45 |
Rasgrp2 |
RAS, guanyl releasing protein 2 |
102 |
0.92 |
| chr17_30591215_30591503 | 0.45 |
Gm50244 |
predicted gene, 50244 |
149 |
0.92 |
| chr16_10395249_10395418 | 0.45 |
Tekt5 |
tektin 5 |
115 |
0.96 |
| chr9_115456234_115456417 | 0.45 |
Gm5921 |
predicted gene 5921 |
17744 |
0.16 |
| chr8_123806538_123806735 | 0.43 |
Rab4a |
RAB4A, member RAS oncogene family |
568 |
0.58 |
| chr19_47304311_47304615 | 0.43 |
Sh3pxd2a |
SH3 and PX domains 2A |
10288 |
0.16 |
| chr18_76117628_76117791 | 0.43 |
2900057B20Rik |
RIKEN cDNA 2900057B20 gene |
19238 |
0.19 |
| chr7_116307697_116308006 | 0.43 |
Plekha7 |
pleckstrin homology domain containing, family A member 7 |
199 |
0.91 |
| chr12_102522079_102522241 | 0.42 |
Chga |
chromogranin A |
32809 |
0.12 |
| chr5_120560470_120560640 | 0.42 |
2510016D11Rik |
RIKEN cDNA 2510016D11 gene |
7516 |
0.1 |
| chr7_98779046_98779213 | 0.42 |
Gm44934 |
predicted gene 44934 |
44048 |
0.1 |
| chr12_57575989_57576174 | 0.41 |
Ttc6 |
tetratricopeptide repeat domain 6 |
264 |
0.9 |
| chr8_117747363_117747696 | 0.41 |
Gm31774 |
predicted gene, 31774 |
731 |
0.61 |
| chr11_101667268_101667443 | 0.40 |
Arl4d |
ADP-ribosylation factor-like 4D |
1814 |
0.19 |
| chr19_26733168_26733326 | 0.40 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
3248 |
0.31 |
| chr2_30966491_30966663 | 0.39 |
Tor1a |
torsin family 1, member A (torsin A) |
1295 |
0.32 |
| chr11_116855946_116856130 | 0.39 |
Mfsd11 |
major facilitator superfamily domain containing 11 |
1960 |
0.2 |
| chr19_8841055_8841224 | 0.38 |
Bscl2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
604 |
0.37 |
| chr5_27677458_27677616 | 0.38 |
Paxip1 |
PAX interacting (with transcription-activation domain) protein 1 |
73758 |
0.1 |
| chr11_60465981_60466150 | 0.38 |
Myo15 |
myosin XV |
3274 |
0.13 |
| chr3_97818935_97819103 | 0.38 |
Pde4dip |
phosphodiesterase 4D interacting protein (myomegalin) |
5602 |
0.23 |
| chr19_4943305_4943466 | 0.38 |
Peli3 |
pellino 3 |
258 |
0.82 |
| chr2_131910128_131910298 | 0.38 |
Prn |
prion protein readthrough transcript |
256 |
0.49 |
| chr4_140001273_140001424 | 0.37 |
Klhdc7a |
kelch domain containing 7A |
33322 |
0.14 |
| chr10_79616142_79616293 | 0.37 |
C2cd4c |
C2 calcium-dependent domain containing 4C |
2192 |
0.15 |
| chr7_67061880_67062042 | 0.36 |
Adamts17 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
14490 |
0.18 |
| chr10_68454663_68454821 | 0.36 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
71025 |
0.1 |
| chr11_113143909_113144540 | 0.36 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
28853 |
0.23 |
| chr11_69605770_69605921 | 0.36 |
Atp1b2 |
ATPase, Na+/K+ transporting, beta 2 polypeptide |
16 |
0.92 |
| chr5_103693522_103693673 | 0.36 |
Aff1 |
AF4/FMR2 family, member 1 |
1223 |
0.43 |
| chr2_154712310_154712496 | 0.36 |
Gm14215 |
predicted gene 14215 |
1319 |
0.32 |
| chr11_78422158_78422337 | 0.35 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
30 |
0.95 |
| chr13_46727661_46727812 | 0.35 |
Nup153 |
nucleoporin 153 |
204 |
0.93 |
| chr19_30145550_30145701 | 0.35 |
Gldc |
glycine decarboxylase |
394 |
0.86 |
| chr11_117810052_117810409 | 0.34 |
Syngr2 |
synaptogyrin 2 |
518 |
0.39 |
| chr15_88866027_88866463 | 0.34 |
Pim3 |
proviral integration site 3 |
1325 |
0.34 |
| chr12_113878948_113879099 | 0.33 |
Ighv2-8 |
immunoglobulin heavy variable V2-8 |
1229 |
0.33 |
| chr4_133049992_133050178 | 0.33 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
10592 |
0.19 |
| chr4_148160555_148160717 | 0.32 |
Fbxo2 |
F-box protein 2 |
15 |
0.65 |
| chr8_84937365_84937533 | 0.32 |
Mast1 |
microtubule associated serine/threonine kinase 1 |
90 |
0.91 |
| chr11_97434945_97435114 | 0.32 |
Arhgap23 |
Rho GTPase activating protein 23 |
1256 |
0.42 |
| chr1_87041212_87041509 | 0.31 |
Gm37447 |
predicted gene, 37447 |
4637 |
0.13 |
| chr11_116175159_116175523 | 0.31 |
Acox1 |
acyl-Coenzyme A oxidase 1, palmitoyl |
5873 |
0.1 |
| chr15_36471981_36472313 | 0.31 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr13_34037312_34037494 | 0.31 |
Bphl |
biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
194 |
0.89 |
| chr14_61680870_61681697 | 0.30 |
Gm37472 |
predicted gene, 37472 |
211 |
0.83 |
| chr2_66256344_66256524 | 0.30 |
Ttc21b |
tetratricopeptide repeat domain 21B |
158 |
0.96 |
| chr12_100343836_100343987 | 0.30 |
Ttc7b |
tetratricopeptide repeat domain 7B |
5615 |
0.19 |
| chr19_6987284_6987440 | 0.30 |
Vegfb |
vascular endothelial growth factor B |
222 |
0.81 |
| chr2_25262755_25263227 | 0.30 |
Tprn |
taperin |
373 |
0.59 |
| chr19_5924482_5924697 | 0.30 |
Cdc42ep2 |
CDC42 effector protein (Rho GTPase binding) 2 |
227 |
0.83 |
| chr13_119713466_119713785 | 0.29 |
Gm48342 |
predicted gene, 48342 |
9076 |
0.11 |
| chr5_37717722_37717888 | 0.29 |
Stk32b |
serine/threonine kinase 32B |
634 |
0.76 |
| chr5_110135214_110135545 | 0.29 |
Chfr |
checkpoint with forkhead and ring finger domains |
463 |
0.65 |
| chr7_39967880_39968059 | 0.29 |
Gm44992 |
predicted gene 44992 |
10105 |
0.24 |
| chr2_155053105_155053256 | 0.29 |
a |
nonagouti |
5565 |
0.16 |
| chr8_12719084_12719404 | 0.28 |
Gm15348 |
predicted gene 15348 |
117 |
0.96 |
| chr4_108217549_108217700 | 0.28 |
Zyg11a |
zyg-11 family member A, cell cycle regulator |
298 |
0.88 |
| chr11_4098038_4098510 | 0.28 |
Mtfp1 |
mitochondrial fission process 1 |
2829 |
0.13 |
| chr14_70051978_70052160 | 0.28 |
Gm33524 |
predicted gene, 33524 |
5298 |
0.19 |
| chr2_167661426_167661577 | 0.28 |
Gm20431 |
predicted gene 20431 |
5 |
0.5 |
| chr11_20826808_20826959 | 0.27 |
Lgalsl |
lectin, galactoside binding-like |
3872 |
0.18 |
| chr2_32681906_32682535 | 0.27 |
Fpgs |
folylpolyglutamyl synthetase |
2784 |
0.1 |
| chr10_88507513_88507686 | 0.27 |
Chpt1 |
choline phosphotransferase 1 |
3526 |
0.19 |
| chr2_156433566_156433753 | 0.27 |
Epb41l1 |
erythrocyte membrane protein band 4.1 like 1 |
12547 |
0.09 |
| chr19_5016908_5017071 | 0.27 |
Slc29a2 |
solute carrier family 29 (nucleoside transporters), member 2 |
6871 |
0.07 |
| chr1_72009620_72009955 | 0.27 |
4933417E11Rik |
RIKEN cDNA 4933417E11 gene |
4592 |
0.18 |
| chr17_12381963_12382316 | 0.27 |
Plg |
plasminogen |
3480 |
0.2 |
| chr2_118111415_118111787 | 0.27 |
Thbs1 |
thrombospondin 1 |
275 |
0.6 |
| chr19_36627251_36627417 | 0.27 |
Hectd2os |
Hectd2, opposite strand |
1310 |
0.49 |
| chr5_111620604_111620778 | 0.26 |
Gm42489 |
predicted gene 42489 |
27075 |
0.18 |
| chr6_113866107_113866258 | 0.26 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
25188 |
0.15 |
| chr1_182620497_182620688 | 0.26 |
Capn8 |
calpain 8 |
55554 |
0.1 |
| chr8_95067717_95067883 | 0.26 |
Drc7 |
dynein regulatory complex subunit 7 |
6279 |
0.11 |
| chr19_8662448_8662612 | 0.26 |
9830166K06Rik |
RIKEN cDNA 9830166K06 gene |
805 |
0.34 |
| chr6_113891432_113891618 | 0.25 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
14 |
0.98 |
| chr12_51471684_51471879 | 0.25 |
Gm18503 |
predicted gene, 18503 |
45545 |
0.14 |
| chr19_4476935_4477185 | 0.25 |
Syt12 |
synaptotagmin XII |
66 |
0.96 |
| chr7_13006960_13007136 | 0.25 |
Zbtb45 |
zinc finger and BTB domain containing 45 |
2752 |
0.11 |
| chr4_138455294_138455445 | 0.25 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
1055 |
0.45 |
| chr11_23124284_23124721 | 0.25 |
1700061J23Rik |
RIKEN cDNA 1700061J23 gene |
27638 |
0.14 |
| chr9_53825348_53825526 | 0.25 |
Slc35f2 |
solute carrier family 35, member F2 |
14720 |
0.17 |
| chr6_118478873_118479106 | 0.25 |
Zfp9 |
zinc finger protein 9 |
331 |
0.85 |
| chr5_137586972_137587155 | 0.25 |
Tfr2 |
transferrin receptor 2 |
3778 |
0.08 |
| chr7_25892258_25892429 | 0.25 |
Cyp2b10 |
cytochrome P450, family 2, subfamily b, polypeptide 10 |
5277 |
0.11 |
| chr14_65785434_65785772 | 0.25 |
Pbk |
PDZ binding kinase |
20234 |
0.18 |
| chr9_103471712_103471904 | 0.25 |
Gm16252 |
predicted gene 16252 |
276 |
0.85 |
| chr17_31631886_31632186 | 0.24 |
Cbs |
cystathionine beta-synthase |
1128 |
0.31 |
| chr13_119713859_119714010 | 0.24 |
Gm48342 |
predicted gene, 48342 |
9385 |
0.11 |
| chr2_118883474_118883639 | 0.24 |
Ivd |
isovaleryl coenzyme A dehydrogenase |
7123 |
0.14 |
| chr5_34636833_34637005 | 0.24 |
Mfsd10 |
major facilitator superfamily domain containing 10 |
78 |
0.95 |
| chr11_87578374_87578544 | 0.24 |
Septin4 |
septin 4 |
3 |
0.97 |
| chr8_84902850_84903029 | 0.24 |
Klf1 |
Kruppel-like factor 1 (erythroid) |
1011 |
0.26 |
| chr2_92383563_92383841 | 0.24 |
1700029I15Rik |
RIKEN cDNA 1700029I15 gene |
784 |
0.45 |
| chr13_117602276_117602441 | 0.24 |
Hcn1 |
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
38 |
0.99 |
| chr12_82333592_82333979 | 0.24 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
8482 |
0.29 |
| chr4_148903004_148903163 | 0.23 |
Casz1 |
castor zinc finger 1 |
1934 |
0.31 |
| chr4_144916418_144916763 | 0.23 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
2289 |
0.32 |
| chr15_101074200_101074369 | 0.23 |
Fignl2 |
fidgetin-like 2 |
4283 |
0.16 |
| chr1_86415976_86416160 | 0.23 |
Nmur1 |
neuromedin U receptor 1 |
10160 |
0.1 |
| chr3_106514922_106515103 | 0.23 |
Dennd2d |
DENN/MADD domain containing 2D |
21837 |
0.11 |
| chr6_121878379_121878709 | 0.23 |
Mug1 |
murinoglobulin 1 |
7033 |
0.2 |
| chr8_68062613_68062915 | 0.23 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
478 |
0.86 |
| chr4_138262391_138262595 | 0.23 |
Kif17 |
kinesin family member 17 |
6 |
0.96 |
| chr9_120933701_120933852 | 0.23 |
Ctnnb1 |
catenin (cadherin associated protein), beta 1 |
42 |
0.95 |
| chr17_7443940_7444098 | 0.22 |
Gm6553 |
predicted gene 6553 |
13777 |
0.19 |
| chr13_55321508_55321662 | 0.22 |
Rab24 |
RAB24, member RAS oncogene family |
194 |
0.71 |
| chr7_130577474_130577931 | 0.22 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
218 |
0.92 |
| chr15_83563229_83563380 | 0.22 |
Tspo |
translocator protein |
288 |
0.63 |
| chr1_45627972_45628123 | 0.22 |
Gm8304 |
predicted gene 8304 |
26021 |
0.17 |
| chr11_95397945_95398308 | 0.22 |
Slc35b1 |
solute carrier family 35, member B1 |
8141 |
0.13 |
| chr5_118004722_118004887 | 0.22 |
Gm42545 |
predicted gene 42545 |
22566 |
0.12 |
| chr15_87045730_87045890 | 0.22 |
Gm23416 |
predicted gene, 23416 |
23183 |
0.18 |
| chr4_122455220_122455373 | 0.22 |
Gm12895 |
predicted gene 12895 |
86 |
0.98 |
| chr2_132869809_132870198 | 0.22 |
Lrrn4 |
leucine rich repeat neuronal 4 |
10888 |
0.13 |
| chr17_28274865_28275039 | 0.21 |
Ppard |
peroxisome proliferator activator receptor delta |
2833 |
0.16 |
| chr15_78178475_78178665 | 0.21 |
Gm49694 |
predicted gene, 49694 |
4109 |
0.16 |
| chr10_80611135_80611333 | 0.21 |
Scamp4 |
secretory carrier membrane protein 4 |
989 |
0.27 |
| chr8_94942711_94942867 | 0.21 |
Adgrg5 |
adhesion G protein-coupled receptor G5 |
8622 |
0.12 |
| chr4_135508053_135508210 | 0.21 |
Gm25317 |
predicted gene, 25317 |
4348 |
0.14 |
| chr10_75160623_75160782 | 0.21 |
Bcr |
BCR activator of RhoGEF and GTPase |
342 |
0.9 |
| chr12_26415244_26415422 | 0.21 |
Rnf144a |
ring finger protein 144A |
79 |
0.79 |
| chr9_21161475_21161850 | 0.21 |
Pde4a |
phosphodiesterase 4A, cAMP specific |
4052 |
0.12 |
| chr16_4268162_4268330 | 0.21 |
Gm5766 |
predicted gene 5766 |
37945 |
0.13 |
| chr8_122660121_122660272 | 0.21 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
13730 |
0.09 |
| chr10_60005449_60005609 | 0.21 |
Ascc1 |
activating signal cointegrator 1 complex subunit 1 |
2202 |
0.26 |
| chr10_97249598_97249782 | 0.21 |
Gm18515 |
predicted gene, 18515 |
7778 |
0.26 |
| chr4_122122855_122123016 | 0.21 |
Gm12896 |
predicted gene 12896 |
88 |
0.98 |
| chr2_109693213_109693439 | 0.21 |
Bdnf |
brain derived neurotrophic factor |
237 |
0.93 |
| chr17_31637083_31637756 | 0.21 |
Cbs |
cystathionine beta-synthase |
181 |
0.89 |
| chr7_129649085_129649245 | 0.21 |
Gm39094 |
predicted gene, 39094 |
9763 |
0.2 |
| chr11_69340671_69341048 | 0.21 |
Rnf227 |
ring finger protein 227 |
50 |
0.93 |
| chr9_123665785_123665936 | 0.21 |
Gm17200 |
predicted gene 17200 |
12236 |
0.09 |
| chr18_46713280_46713447 | 0.20 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
14666 |
0.13 |
| chr18_35662151_35662309 | 0.20 |
Spata24 |
spermatogenesis associated 24 |
44 |
0.51 |
| chr11_74638791_74638959 | 0.20 |
Cluh |
clustered mitochondria (cluA/CLU1) homolog |
10620 |
0.15 |
| chr12_102623965_102624273 | 0.20 |
Gm20069 |
predicted gene, 20069 |
11439 |
0.13 |
| chr13_60479839_60480430 | 0.20 |
Gm48500 |
predicted gene, 48500 |
1164 |
0.45 |
| chr6_42325328_42325493 | 0.20 |
Fam131b |
family with sequence similarity 131, member B |
767 |
0.44 |
| chr14_22019673_22019870 | 0.20 |
Lrmda |
leucine rich melanocyte differentiation associated |
50 |
0.96 |
| chr1_92647884_92648048 | 0.20 |
Otos |
otospiralin |
288 |
0.83 |
| chr2_152081883_152082034 | 0.20 |
Scrt2 |
scratch family zinc finger 2 |
429 |
0.78 |
| chr9_103258204_103258373 | 0.20 |
1300017J02Rik |
RIKEN cDNA 1300017J02 gene |
1262 |
0.42 |
| chr5_125271875_125272026 | 0.20 |
Gm32585 |
predicted gene, 32585 |
865 |
0.57 |
| chr17_31390196_31390347 | 0.20 |
Pde9a |
phosphodiesterase 9A |
3977 |
0.17 |
| chr14_20794215_20794517 | 0.20 |
Gm30108 |
predicted gene, 30108 |
127 |
0.69 |
| chr19_28975451_28975625 | 0.20 |
Gm3621 |
predicted gene 3621 |
2499 |
0.17 |
| chr6_128736252_128736414 | 0.20 |
Gm43907 |
predicted gene, 43907 |
8311 |
0.11 |
| chr1_180991444_180991626 | 0.20 |
Ephx1 |
epoxide hydrolase 1, microsomal |
4635 |
0.11 |
| chrX_101419042_101419230 | 0.20 |
Zmym3 |
zinc finger, MYM-type 3 |
662 |
0.61 |
| chr15_102126408_102126761 | 0.20 |
Spryd3 |
SPRY domain containing 3 |
2300 |
0.17 |
| chr11_50431056_50431230 | 0.20 |
Rufy1 |
RUN and FYVE domain containing 1 |
18 |
0.98 |
| chr10_70599538_70599732 | 0.19 |
Phyhipl |
phytanoyl-CoA hydroxylase interacting protein-like |
344 |
0.91 |
| chr4_116008423_116008814 | 0.19 |
Faah |
fatty acid amide hydrolase |
8550 |
0.14 |
| chr13_7428915_7429066 | 0.19 |
Gm36074 |
predicted gene, 36074 |
17519 |
0.27 |
| chr17_56935189_56935340 | 0.19 |
Mllt1 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 1 |
151 |
0.92 |
| chr9_30124659_30124991 | 0.19 |
Gm47677 |
predicted gene, 47677 |
42750 |
0.18 |
| chr10_125949154_125949327 | 0.19 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
16928 |
0.28 |
| chr9_35139662_35139830 | 0.19 |
Gm24262 |
predicted gene, 24262 |
4890 |
0.14 |
| chr12_113859003_113859160 | 0.19 |
Ighv5-18 |
immunoglobulin heavy variable V5-18 |
17158 |
0.11 |
| chr16_85173682_85173850 | 0.19 |
App |
amyloid beta (A4) precursor protein |
0 |
0.98 |
| chr6_147042911_147043077 | 0.19 |
Mrps35 |
mitochondrial ribosomal protein S35 |
230 |
0.89 |
| chr15_76369272_76369423 | 0.19 |
Hgh1 |
HGH1 homolog |
134 |
0.88 |
| chr2_179290486_179290655 | 0.19 |
Gm14293 |
predicted gene 14293 |
50080 |
0.15 |
| chr16_20537030_20537361 | 0.19 |
Ap2m1 |
adaptor-related protein complex 2, mu 1 subunit |
417 |
0.62 |
| chr1_74338040_74338254 | 0.19 |
Pnkd |
paroxysmal nonkinesiogenic dyskinesia |
5484 |
0.1 |
| chr15_81849441_81849616 | 0.19 |
Gm8444 |
predicted gene 8444 |
5815 |
0.1 |
| chr11_94732887_94733059 | 0.19 |
Cdc34b |
cell division cycle 34B |
8809 |
0.11 |
| chr11_102753302_102753453 | 0.19 |
Adam11 |
a disintegrin and metallopeptidase domain 11 |
8062 |
0.11 |
| chr18_5085122_5085283 | 0.19 |
Svil |
supervillin |
9179 |
0.28 |
| chr3_89436568_89436763 | 0.19 |
Pbxip1 |
pre B cell leukemia transcription factor interacting protein 1 |
41 |
0.94 |
| chr8_20557902_20558131 | 0.19 |
Gm21182 |
predicted gene, 21182 |
530 |
0.76 |
| chr3_107631354_107631565 | 0.19 |
Gm10961 |
predicted gene 10961 |
137 |
0.63 |
| chr5_66618891_66619162 | 0.18 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
242 |
0.91 |
| chr15_36609144_36609341 | 0.18 |
Pabpc1 |
poly(A) binding protein, cytoplasmic 1 |
269 |
0.88 |
| chr2_164952558_164952709 | 0.18 |
Mmp9 |
matrix metallopeptidase 9 |
1753 |
0.24 |
| chr15_83724999_83725153 | 0.18 |
Scube1 |
signal peptide, CUB domain, EGF-like 1 |
55 |
0.98 |
| chr1_181475618_181475786 | 0.18 |
Ccdc121 |
coiled-coil domain containing 121 |
35749 |
0.14 |
| chr5_121546456_121546786 | 0.18 |
Mapkapk5 |
MAP kinase-activated protein kinase 5 |
716 |
0.35 |
| chr18_34373553_34373724 | 0.18 |
Reep5 |
receptor accessory protein 5 |
269 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.4 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.3 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.6 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.1 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.1 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.2 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.2 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.2 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.1 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.2 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.1 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.3 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.4 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.0 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.0 | GO:0045404 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) regulation of eosinophil activation(GO:1902566) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.0 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.2 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.0 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.0 | GO:0060375 | mast cell differentiation(GO:0060374) regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.0 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:1901679 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0000938 | GARP complex(GO:0000938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.2 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.2 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.4 | GO:0052677 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.0 | REACTOME PLATELET HOMEOSTASIS | Genes involved in Platelet homeostasis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |