Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
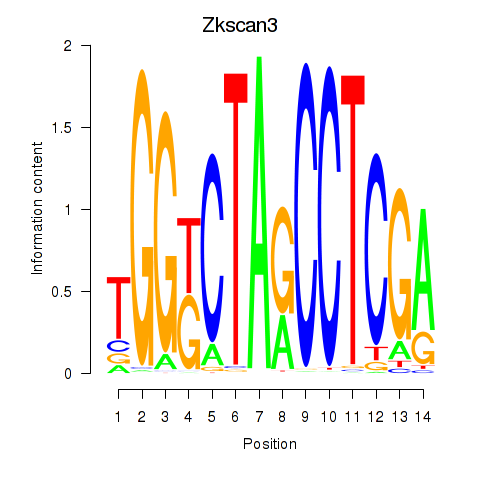
Results for Zkscan3
Z-value: 3.04
Transcription factors associated with Zkscan3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan3
|
ENSMUSG00000021327.12 | zinc finger with KRAB and SCAN domains 3 |
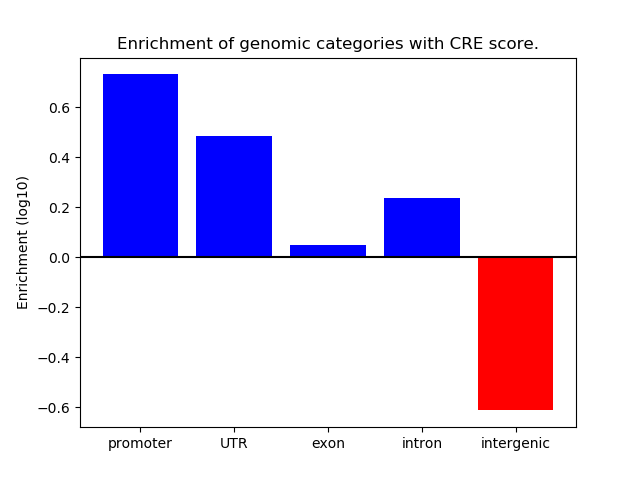
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_21402344_21402506 | Zkscan3 | 245 | 0.807795 | 0.75 | 8.8e-02 | Click! |
| chr13_21402673_21402832 | Zkscan3 | 3 | 0.938289 | 0.60 | 2.1e-01 | Click! |
| chr13_21401689_21401902 | Zkscan3 | 875 | 0.336824 | 0.58 | 2.2e-01 | Click! |
| chr13_21402012_21402170 | Zkscan3 | 579 | 0.506873 | 0.52 | 2.9e-01 | Click! |
| chr13_21401499_21401673 | Zkscan3 | 1084 | 0.264744 | -0.35 | 5.0e-01 | Click! |
Activity of the Zkscan3 motif across conditions
Conditions sorted by the z-value of the Zkscan3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
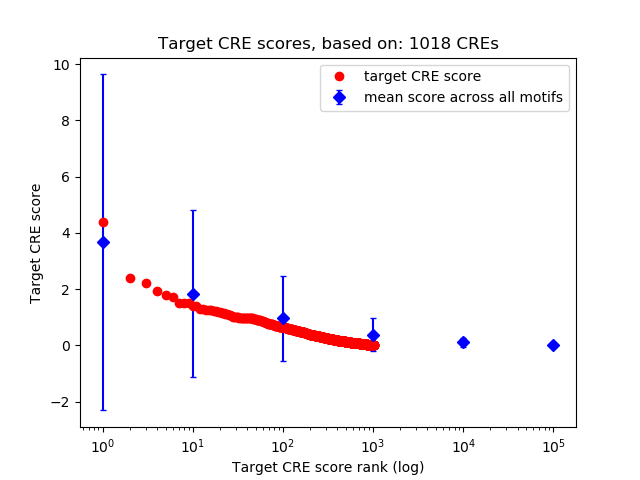
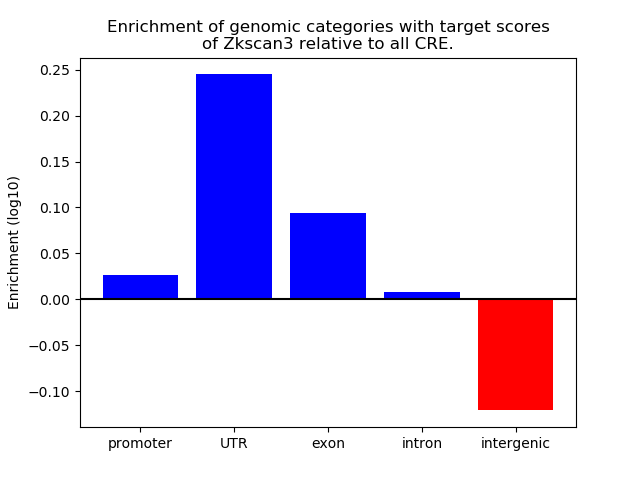
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_65334133_65334359 | 4.38 |
Gm39363 |
predicted gene, 39363 |
1726 |
0.18 |
| chr8_35387028_35387898 | 2.40 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
10803 |
0.16 |
| chr13_93628549_93628834 | 2.21 |
Gm15622 |
predicted gene 15622 |
3309 |
0.2 |
| chr16_97900645_97900797 | 1.94 |
C2cd2 |
C2 calcium-dependent domain containing 2 |
8440 |
0.18 |
| chr5_112299397_112299785 | 1.79 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
4305 |
0.13 |
| chr7_98352590_98353259 | 1.71 |
Tsku |
tsukushi, small leucine rich proteoglycan |
7155 |
0.18 |
| chr6_122819078_122819314 | 1.52 |
Foxj2 |
forkhead box J2 |
718 |
0.44 |
| chr9_61826654_61826867 | 1.52 |
Gm19208 |
predicted gene, 19208 |
24944 |
0.2 |
| chr11_4906060_4906238 | 1.50 |
Thoc5 |
THO complex 5 |
3937 |
0.15 |
| chr3_98226583_98226734 | 1.40 |
Reg4 |
regenerating islet-derived family, member 4 |
4502 |
0.17 |
| chr6_144781221_144781581 | 1.40 |
Sox5 |
SRY (sex determining region Y)-box 5 |
518 |
0.79 |
| chr14_21171384_21171535 | 1.30 |
Adk |
adenosine kinase |
95307 |
0.07 |
| chr19_3650199_3650464 | 1.28 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
34539 |
0.1 |
| chr14_76556939_76557219 | 1.27 |
Serp2 |
stress-associated endoplasmic reticulum protein family member 2 |
190 |
0.95 |
| chr17_28436590_28436749 | 1.26 |
Fkbp5 |
FK506 binding protein 5 |
4341 |
0.12 |
| chr19_61048296_61048486 | 1.24 |
Gm22520 |
predicted gene, 22520 |
34846 |
0.14 |
| chr7_125578941_125579144 | 1.21 |
Gm44876 |
predicted gene 44876 |
6925 |
0.16 |
| chr8_120369815_120369966 | 1.21 |
Gm22715 |
predicted gene, 22715 |
73659 |
0.09 |
| chr17_33932399_33932565 | 1.20 |
Rgl2 |
ral guanine nucleotide dissociation stimulator-like 2 |
351 |
0.58 |
| chr8_41026178_41026329 | 1.17 |
Mtus1 |
mitochondrial tumor suppressor 1 |
3351 |
0.19 |
| chr7_98849207_98849358 | 1.14 |
Wnt11 |
wingless-type MMTV integration site family, member 11 |
1476 |
0.34 |
| chr10_95317812_95318192 | 1.13 |
Gm48882 |
predicted gene, 48882 |
245 |
0.88 |
| chr2_153586432_153586583 | 1.13 |
Commd7 |
COMM domain containing 7 |
46224 |
0.11 |
| chr1_74061177_74061331 | 1.13 |
Tns1 |
tensin 1 |
24097 |
0.17 |
| chr6_145764862_145765013 | 1.09 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
14366 |
0.19 |
| chr14_100324093_100324244 | 1.07 |
Gm41231 |
predicted gene, 41231 |
38003 |
0.15 |
| chr15_36472644_36473037 | 1.06 |
Ankrd46 |
ankyrin repeat domain 46 |
23875 |
0.13 |
| chr10_80340392_80340718 | 1.02 |
Adamtsl5 |
ADAMTS-like 5 |
470 |
0.56 |
| chr8_124201642_124201985 | 1.00 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
29578 |
0.15 |
| chr4_140858134_140858561 | 1.00 |
Padi1 |
peptidyl arginine deiminase, type I |
12569 |
0.12 |
| chr10_122986042_122986255 | 1.00 |
D630033A02Rik |
RIKEN cDNA D630033A02 gene |
317 |
0.58 |
| chr18_73844340_73844698 | 0.99 |
Mro |
maestro |
14866 |
0.2 |
| chr17_46110701_46110892 | 0.99 |
Mrps18a |
mitochondrial ribosomal protein S18A |
190 |
0.91 |
| chr5_126543958_126544274 | 0.99 |
Gm24839 |
predicted gene, 24839 |
30135 |
0.19 |
| chr9_115476457_115476622 | 0.99 |
Gm5921 |
predicted gene 5921 |
37958 |
0.14 |
| chr6_42352219_42352377 | 0.98 |
Zyx |
zyxin |
1180 |
0.24 |
| chr18_70568874_70569217 | 0.98 |
Mbd2 |
methyl-CpG binding domain protein 2 |
711 |
0.68 |
| chr2_167326193_167326424 | 0.98 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
12408 |
0.18 |
| chr2_126570201_126570813 | 0.98 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
5747 |
0.2 |
| chr17_30619245_30619423 | 0.97 |
Dnah8 |
dynein, axonemal, heavy chain 8 |
5020 |
0.12 |
| chr7_127214965_127215278 | 0.97 |
Septin1 |
septin 1 |
4 |
0.92 |
| chr5_24586259_24586557 | 0.97 |
Chpf2 |
chondroitin polymerizing factor 2 |
333 |
0.76 |
| chr9_118925675_118926262 | 0.97 |
Gm2415 |
predicted gene 2415 |
290 |
0.62 |
| chr13_19395072_19395660 | 0.96 |
Gm42683 |
predicted gene 42683 |
87 |
0.82 |
| chr9_32004053_32004204 | 0.96 |
Gm47465 |
predicted gene, 47465 |
1332 |
0.4 |
| chr8_124897995_124898160 | 0.96 |
Sprtn |
SprT-like N-terminal domain |
191 |
0.68 |
| chr4_149978768_149979375 | 0.95 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
22194 |
0.13 |
| chr13_28421437_28421636 | 0.94 |
Gm40841 |
predicted gene, 40841 |
1470 |
0.49 |
| chr16_33947299_33947485 | 0.94 |
Itgb5 |
integrin beta 5 |
868 |
0.56 |
| chr4_133049992_133050178 | 0.92 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
10592 |
0.19 |
| chr15_79141450_79141641 | 0.92 |
Polr2f |
polymerase (RNA) II (DNA directed) polypeptide F |
179 |
0.62 |
| chr4_118132718_118132924 | 0.91 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
2061 |
0.28 |
| chr3_103789902_103790723 | 0.90 |
Hipk1 |
homeodomain interacting protein kinase 1 |
763 |
0.4 |
| chr3_101519636_101519818 | 0.90 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
57833 |
0.1 |
| chr9_70940473_70940653 | 0.88 |
Lipc |
lipase, hepatic |
5755 |
0.22 |
| chr13_113864739_113864979 | 0.88 |
Arl15 |
ADP-ribosylation factor-like 15 |
70237 |
0.09 |
| chr8_12677440_12677591 | 0.88 |
4931415C17Rik |
RIKEN cDNA 4931415C17 gene |
5088 |
0.15 |
| chr13_60644455_60644612 | 0.87 |
Gm48583 |
predicted gene, 48583 |
2743 |
0.25 |
| chr15_34469803_34469957 | 0.86 |
Gm49188 |
predicted gene, 49188 |
11827 |
0.1 |
| chr12_100199636_100199926 | 0.86 |
Calm1 |
calmodulin 1 |
252 |
0.89 |
| chr2_75603496_75603684 | 0.84 |
Gm13655 |
predicted gene 13655 |
29792 |
0.14 |
| chr11_116104363_116104575 | 0.82 |
Trim47 |
tripartite motif-containing 47 |
2614 |
0.14 |
| chr2_30268240_30268600 | 0.82 |
Phyhd1 |
phytanoyl-CoA dioxygenase domain containing 1 |
1557 |
0.21 |
| chr10_81419362_81419546 | 0.81 |
Mir1191b |
microRNA 1191b |
3157 |
0.08 |
| chr19_7240751_7240928 | 0.80 |
Naa40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
195 |
0.89 |
| chr5_124353375_124353533 | 0.80 |
Cdk2ap1 |
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
1217 |
0.33 |
| chr5_89462978_89463338 | 0.79 |
Gc |
vitamin D binding protein |
5260 |
0.24 |
| chr2_32682708_32682915 | 0.78 |
Fpgs |
folylpolyglutamyl synthetase |
2193 |
0.12 |
| chr8_11262573_11262724 | 0.77 |
Col4a1 |
collagen, type IV, alpha 1 |
17008 |
0.16 |
| chr19_5851015_5851290 | 0.77 |
Frmd8os |
FERM domain containing 8, opposite strand |
2644 |
0.11 |
| chr16_17235100_17235306 | 0.77 |
Hic2 |
hypermethylated in cancer 2 |
1539 |
0.2 |
| chr9_96727975_96728455 | 0.76 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
821 |
0.58 |
| chr18_20670501_20671004 | 0.76 |
Ttr |
transthyretin |
5472 |
0.18 |
| chr3_137863947_137864380 | 0.76 |
Gm43403 |
predicted gene 43403 |
20 |
0.81 |
| chr19_44400900_44401269 | 0.76 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5606 |
0.16 |
| chr7_16597600_16597898 | 0.75 |
Gm29443 |
predicted gene 29443 |
16075 |
0.09 |
| chr9_90109465_90109757 | 0.74 |
Morf4l1 |
mortality factor 4 like 1 |
4678 |
0.17 |
| chr16_38433421_38433613 | 0.74 |
Pla1a |
phospholipase A1 member A |
372 |
0.8 |
| chr11_60786856_60787068 | 0.73 |
Shmt1 |
serine hydroxymethyltransferase 1 (soluble) |
6211 |
0.09 |
| chr19_46908820_46909200 | 0.73 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
565 |
0.74 |
| chr13_54076591_54076742 | 0.71 |
Sfxn1 |
sideroflexin 1 |
4797 |
0.21 |
| chr2_30436208_30436447 | 0.70 |
Ptpa |
protein phosphatase 2 protein activator |
1902 |
0.22 |
| chr3_146403874_146404213 | 0.70 |
Ssx2ip |
synovial sarcoma, X 2 interacting protein |
599 |
0.64 |
| chr7_98177945_98178149 | 0.69 |
Capn5 |
calpain 5 |
227 |
0.83 |
| chr11_75422344_75422535 | 0.69 |
Serpinf1 |
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
113 |
0.88 |
| chr2_25708935_25709097 | 0.68 |
A230005M16Rik |
RIKEN cDNA A230005M16 gene |
700 |
0.42 |
| chr17_47873312_47873495 | 0.68 |
Foxp4 |
forkhead box P4 |
5550 |
0.14 |
| chr6_42251736_42251903 | 0.68 |
Gstk1 |
glutathione S-transferase kappa 1 |
5796 |
0.11 |
| chr2_173156797_173157495 | 0.68 |
Pck1 |
phosphoenolpyruvate carboxykinase 1, cytosolic |
4064 |
0.2 |
| chr9_48738331_48738489 | 0.67 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
97535 |
0.07 |
| chr4_62779675_62780043 | 0.67 |
Gm24117 |
predicted gene, 24117 |
32569 |
0.15 |
| chr2_32421538_32422351 | 0.67 |
Slc25a25 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
36 |
0.95 |
| chr16_84805927_84806078 | 0.66 |
Jam2 |
junction adhesion molecule 2 |
3405 |
0.17 |
| chr14_66129671_66129842 | 0.66 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
5256 |
0.17 |
| chr4_95625117_95625268 | 0.65 |
Fggy |
FGGY carbohydrate kinase domain containing |
1636 |
0.46 |
| chr9_122049922_122050484 | 0.65 |
Gm39465 |
predicted gene, 39465 |
1155 |
0.34 |
| chr9_20740746_20740928 | 0.65 |
Olfm2 |
olfactomedin 2 |
5512 |
0.16 |
| chr2_118844371_118844522 | 0.65 |
Gm14091 |
predicted gene 14091 |
10324 |
0.12 |
| chr6_113644583_113645202 | 0.65 |
Gm43964 |
predicted gene, 43964 |
3371 |
0.09 |
| chr11_102267338_102267489 | 0.65 |
Asb16 |
ankyrin repeat and SOCS box-containing 16 |
1330 |
0.26 |
| chr2_94067079_94067259 | 0.64 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
405 |
0.86 |
| chr19_20450928_20451527 | 0.64 |
C730002L08Rik |
RIKEN cDNA C730002L08 gene |
26445 |
0.16 |
| chr10_81233301_81233523 | 0.64 |
Zfr2 |
zinc finger RNA binding protein 2 |
225 |
0.8 |
| chr4_116006006_116006178 | 0.64 |
Faah |
fatty acid amide hydrolase |
6024 |
0.15 |
| chr11_75211079_75211230 | 0.63 |
Rtn4rl1 |
reticulon 4 receptor-like 1 |
17371 |
0.1 |
| chr10_24831746_24832070 | 0.63 |
Enpp3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
102 |
0.97 |
| chr11_107025954_107026367 | 0.63 |
1810010H24Rik |
RIKEN cDNA 1810010H24 gene |
479 |
0.74 |
| chr2_167328678_167328847 | 0.62 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
14862 |
0.18 |
| chr15_99032417_99032770 | 0.62 |
Tuba1c |
tubulin, alpha 1C |
2272 |
0.15 |
| chr15_76745135_76745286 | 0.62 |
Arhgap39 |
Rho GTPase activating protein 39 |
19607 |
0.08 |
| chr15_31264328_31264561 | 0.62 |
Dap |
death-associated protein |
3892 |
0.19 |
| chr16_93823192_93823406 | 0.61 |
Morc3 |
microrchidia 3 |
8822 |
0.13 |
| chr10_33950641_33951001 | 0.60 |
Zup1 |
zinc finger containing ubiquitin peptidase 1 |
374 |
0.76 |
| chr9_53301822_53302322 | 0.60 |
Exph5 |
exophilin 5 |
402 |
0.86 |
| chr9_70234355_70234506 | 0.60 |
Myo1e |
myosin IE |
27062 |
0.19 |
| chr19_61048615_61048813 | 0.59 |
Gm22520 |
predicted gene, 22520 |
35169 |
0.14 |
| chr17_45834393_45834544 | 0.59 |
Gm35692 |
predicted gene, 35692 |
12145 |
0.16 |
| chr18_75601004_75601199 | 0.59 |
Ctif |
CBP80/20-dependent translation initiation factor |
36172 |
0.19 |
| chr19_43510545_43510713 | 0.58 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
5236 |
0.13 |
| chrX_75874057_75874225 | 0.58 |
Pls3 |
plastin 3 (T-isoform) |
433 |
0.81 |
| chr6_21828351_21828787 | 0.58 |
Tspan12 |
tetraspanin 12 |
23259 |
0.17 |
| chr14_70198497_70198662 | 0.57 |
Sorbs3 |
sorbin and SH3 domain containing 3 |
6044 |
0.12 |
| chr16_37635141_37635534 | 0.57 |
Hgd |
homogentisate 1, 2-dioxygenase |
13488 |
0.15 |
| chr5_66148784_66148935 | 0.57 |
Rbm47 |
RNA binding motif protein 47 |
2097 |
0.18 |
| chr1_88286437_88286600 | 0.57 |
Trpm8 |
transient receptor potential cation channel, subfamily M, member 8 |
8857 |
0.11 |
| chr8_124567905_124568437 | 0.56 |
Agt |
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
1535 |
0.36 |
| chr5_52975314_52975474 | 0.56 |
Gm30301 |
predicted gene, 30301 |
6643 |
0.16 |
| chr15_65846454_65846635 | 0.56 |
Efr3a |
EFR3 homolog A |
4577 |
0.22 |
| chr14_45220729_45220892 | 0.55 |
Txndc16 |
thioredoxin domain containing 16 |
482 |
0.52 |
| chr10_70097931_70098284 | 0.55 |
Ccdc6 |
coiled-coil domain containing 6 |
986 |
0.66 |
| chr8_120366684_120367118 | 0.55 |
Gm22715 |
predicted gene, 22715 |
76648 |
0.08 |
| chr9_62342926_62343110 | 0.55 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
554 |
0.78 |
| chr2_31479577_31479870 | 0.54 |
Ass1 |
argininosuccinate synthetase 1 |
9516 |
0.19 |
| chr9_62367194_62367386 | 0.54 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
346 |
0.89 |
| chr4_132628872_132629046 | 0.53 |
Eya3 |
EYA transcriptional coactivator and phosphatase 3 |
10028 |
0.15 |
| chr15_79733610_79733958 | 0.53 |
Sun2 |
Sad1 and UNC84 domain containing 2 |
2167 |
0.15 |
| chr16_13201906_13202058 | 0.53 |
Mrtfb |
myocardin related transcription factor B |
54499 |
0.14 |
| chr16_72830483_72830654 | 0.53 |
Robo1 |
roundabout guidance receptor 1 |
145781 |
0.05 |
| chr7_79831481_79831780 | 0.52 |
Gm44974 |
predicted gene 44974 |
241 |
0.87 |
| chr16_85901352_85901509 | 0.52 |
Adamts5 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) |
398 |
0.9 |
| chr2_163301297_163301733 | 0.52 |
Tox2 |
TOX high mobility group box family member 2 |
18863 |
0.19 |
| chr10_61143381_61143586 | 0.52 |
Sgpl1 |
sphingosine phosphate lyase 1 |
3415 |
0.19 |
| chr16_45975826_45976145 | 0.52 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
22387 |
0.14 |
| chr5_73337973_73338415 | 0.52 |
Ociad2 |
OCIA domain containing 2 |
364 |
0.79 |
| chr19_6855220_6855477 | 0.52 |
Ccdc88b |
coiled-coil domain containing 88B |
110 |
0.93 |
| chr1_86879397_86879607 | 0.51 |
Gm37541 |
predicted gene, 37541 |
5152 |
0.12 |
| chr16_91833643_91834213 | 0.50 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
5771 |
0.19 |
| chr6_91655124_91655309 | 0.50 |
Gm45217 |
predicted gene 45217 |
24007 |
0.11 |
| chr8_33871610_33871761 | 0.50 |
Gm26978 |
predicted gene, 26978 |
14062 |
0.16 |
| chr1_158142001_158142160 | 0.50 |
Brinp2 |
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
154384 |
0.04 |
| chr9_106756626_106756783 | 0.50 |
Rad54l2 |
RAD54 like 2 (S. cerevisiae) |
32472 |
0.12 |
| chr8_119912901_119913087 | 0.49 |
Usp10 |
ubiquitin specific peptidase 10 |
2137 |
0.24 |
| chr7_79781946_79782169 | 0.49 |
Gm44705 |
predicted gene 44705 |
8568 |
0.1 |
| chr10_81422057_81422238 | 0.49 |
Nfic |
nuclear factor I/C |
4967 |
0.07 |
| chr13_99099705_99099869 | 0.49 |
Gm807 |
predicted gene 807 |
919 |
0.55 |
| chr4_43421375_43421673 | 0.49 |
Rusc2 |
RUN and SH3 domain containing 2 |
968 |
0.4 |
| chr9_44387040_44387229 | 0.49 |
Hyou1 |
hypoxia up-regulated 1 |
3527 |
0.07 |
| chr4_8754704_8754856 | 0.49 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
3342 |
0.36 |
| chr2_180906861_180907031 | 0.48 |
Gm27032 |
predicted gene, 27032 |
12841 |
0.09 |
| chr17_71255942_71256093 | 0.48 |
Emilin2 |
elastin microfibril interfacer 2 |
329 |
0.86 |
| chr13_37874544_37874704 | 0.48 |
Rreb1 |
ras responsive element binding protein 1 |
13810 |
0.2 |
| chr19_6924075_6924226 | 0.48 |
Catsperz |
cation channel sperm associated auxiliary subunit zeta |
719 |
0.37 |
| chr8_83589428_83589579 | 0.48 |
Gm45823 |
predicted gene 45823 |
3978 |
0.11 |
| chrX_140525251_140525408 | 0.48 |
Tsc22d3 |
TSC22 domain family, member 3 |
17339 |
0.19 |
| chr8_35207158_35207309 | 0.47 |
Gm34474 |
predicted gene, 34474 |
11405 |
0.14 |
| chr10_111364892_111365156 | 0.47 |
Gm40761 |
predicted gene, 40761 |
27900 |
0.17 |
| chr9_14439768_14439948 | 0.47 |
Gm47318 |
predicted gene, 47318 |
4447 |
0.13 |
| chr10_75794436_75794892 | 0.47 |
Gstt1 |
glutathione S-transferase, theta 1 |
2905 |
0.13 |
| chr8_81378490_81378647 | 0.46 |
Inpp4b |
inositol polyphosphate-4-phosphatase, type II |
36006 |
0.19 |
| chr8_80856636_80856789 | 0.46 |
Gm31105 |
predicted gene, 31105 |
23228 |
0.16 |
| chr10_69102086_69102245 | 0.46 |
Gm47107 |
predicted gene, 47107 |
4299 |
0.21 |
| chr3_52605604_52605772 | 0.46 |
Gm10293 |
predicted pseudogene 10293 |
7147 |
0.26 |
| chr7_116443673_116444404 | 0.45 |
AV356131 |
expressed sequence AV356131 |
463 |
0.52 |
| chr7_30895336_30895491 | 0.45 |
D7Ertd128e |
DNA segment, Chr 7, ERATO Doi 128, expressed |
10382 |
0.07 |
| chr19_58021843_58021994 | 0.45 |
Mir5623 |
microRNA 5623 |
29249 |
0.23 |
| chr19_5831533_5832364 | 0.45 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
3936 |
0.09 |
| chr7_123407139_123407415 | 0.45 |
Lcmt1 |
leucine carboxyl methyltransferase 1 |
903 |
0.58 |
| chr8_24457283_24457668 | 0.44 |
Gm44620 |
predicted gene 44620 |
10378 |
0.15 |
| chr7_3331834_3332020 | 0.44 |
Cacng7 |
calcium channel, voltage-dependent, gamma subunit 7 |
1028 |
0.31 |
| chr9_98986714_98987086 | 0.44 |
Faim |
Fas apoptotic inhibitory molecule |
493 |
0.73 |
| chr15_83452182_83452447 | 0.44 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
12238 |
0.15 |
| chr1_60609032_60609279 | 0.44 |
Gm23762 |
predicted gene, 23762 |
4648 |
0.15 |
| chr9_21725120_21725502 | 0.43 |
Ldlr |
low density lipoprotein receptor |
1670 |
0.23 |
| chr6_28737275_28737426 | 0.43 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
27340 |
0.19 |
| chr9_102737824_102737975 | 0.42 |
Gm5627 |
predicted gene 5627 |
1749 |
0.25 |
| chr18_80254812_80254963 | 0.42 |
Slc66a2 |
solute carrier family 66 member 2 |
358 |
0.79 |
| chr10_68259248_68259399 | 0.41 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
19398 |
0.21 |
| chr4_152027330_152027509 | 0.41 |
Zbtb48 |
zinc finger and BTB domain containing 48 |
218 |
0.87 |
| chr7_114769396_114769572 | 0.41 |
Insc |
INSC spindle orientation adaptor protein |
710 |
0.68 |
| chr13_43499439_43499647 | 0.41 |
Gm47683 |
predicted gene, 47683 |
624 |
0.64 |
| chr7_119941261_119941568 | 0.41 |
Lyrm1 |
LYR motif containing 1 |
4456 |
0.17 |
| chr17_12921475_12921639 | 0.40 |
Snora20 |
small nucleolar RNA, H/ACA box 20 |
1233 |
0.19 |
| chr7_19457920_19458071 | 0.40 |
Mark4 |
MAP/microtubule affinity regulating kinase 4 |
800 |
0.36 |
| chr2_158070723_158070992 | 0.40 |
2010009K17Rik |
RIKEN cDNA 2010009K17 gene |
20940 |
0.14 |
| chr9_80067924_80068106 | 0.40 |
Senp6 |
SUMO/sentrin specific peptidase 6 |
198 |
0.93 |
| chr18_54984622_54984821 | 0.39 |
Zfp608 |
zinc finger protein 608 |
5445 |
0.22 |
| chr2_27696621_27697049 | 0.39 |
Rxra |
retinoid X receptor alpha |
12457 |
0.25 |
| chr19_46627552_46627707 | 0.39 |
Wbp1l |
WW domain binding protein 1 like |
4228 |
0.16 |
| chr11_100921491_100921782 | 0.38 |
Stat3 |
signal transducer and activator of transcription 3 |
13412 |
0.14 |
| chr9_69290695_69290919 | 0.38 |
Rora |
RAR-related orphan receptor alpha |
1125 |
0.59 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.4 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.1 | 0.8 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.3 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.5 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.3 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 0.3 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.2 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.2 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.6 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.3 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:1903935 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) response to sodium arsenite(GO:1903935) |
| 0.0 | 0.1 | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0060921 | sinoatrial node cell differentiation(GO:0060921) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.5 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.2 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0061623 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.8 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.5 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.0 | GO:0045297 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.0 | GO:0021843 | substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.0 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.0 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.6 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.0 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 1.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 0.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 0.6 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.5 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.3 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.3 | GO:0052811 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.7 | GO:0018711 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |