Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
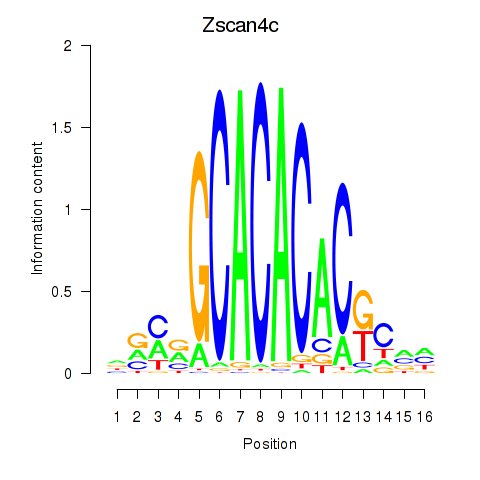
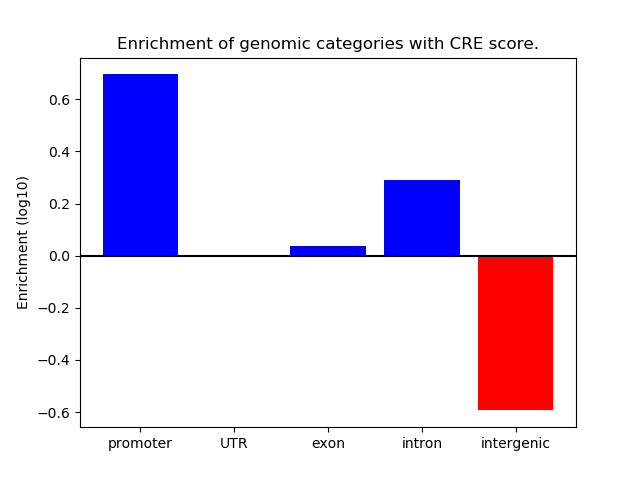
Results for Zscan4c
Z-value: 1.77
Transcription factors associated with Zscan4c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zscan4c
|
ENSMUSG00000054272.5 | zinc finger and SCAN domain containing 4C |
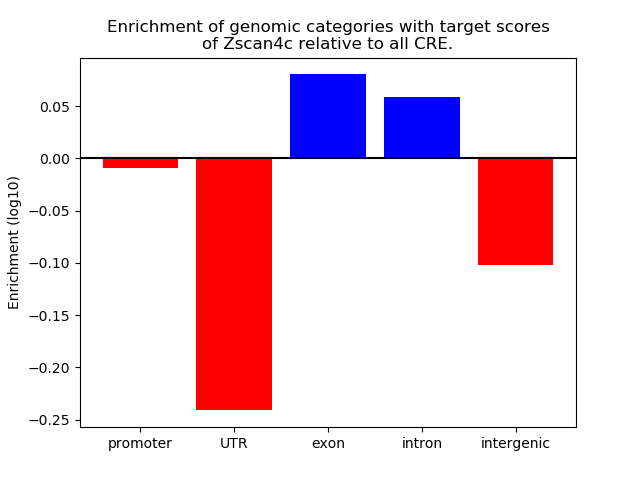
Activity of the Zscan4c motif across conditions
Conditions sorted by the z-value of the Zscan4c motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
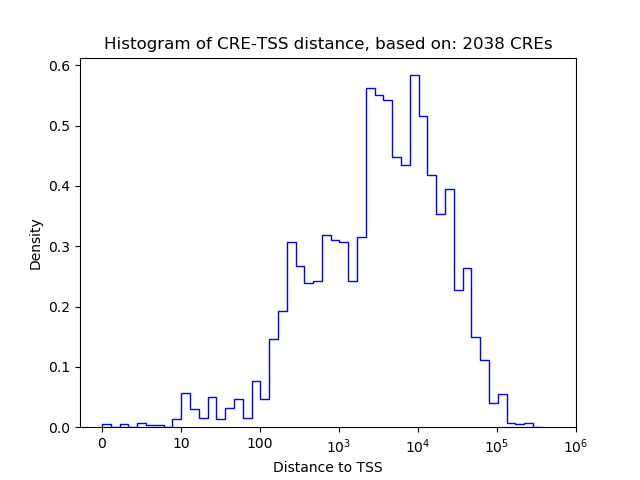
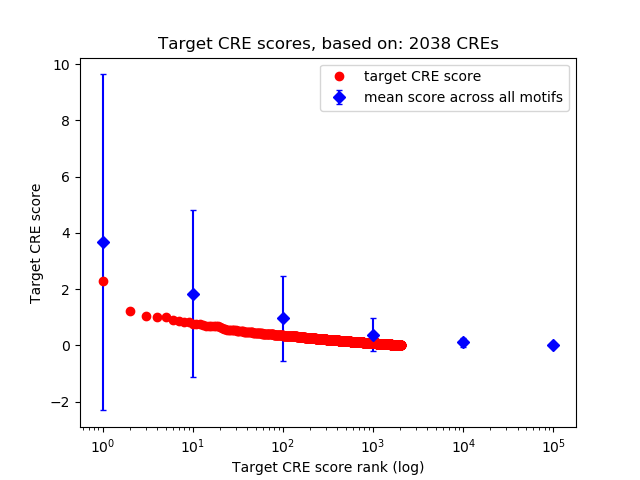
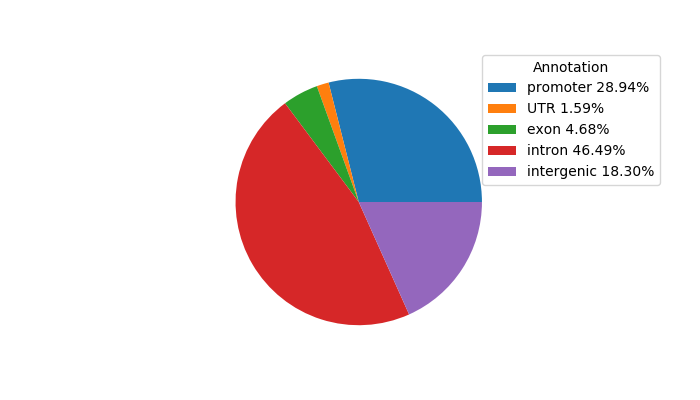
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_12732086_12732318 | 2.30 |
Ttc39c |
tetratricopeptide repeat domain 39C |
2503 |
0.2 |
| chr18_12731812_12732024 | 1.23 |
Ttc39c |
tetratricopeptide repeat domain 39C |
2787 |
0.19 |
| chr5_38117047_38117323 | 1.04 |
Stx18 |
syntaxin 18 |
3724 |
0.2 |
| chr9_122119702_122119865 | 1.02 |
Gm47122 |
predicted gene, 47122 |
638 |
0.56 |
| chr1_162966082_162966252 | 0.99 |
Gm37273 |
predicted gene, 37273 |
16847 |
0.15 |
| chr11_117831026_117831177 | 0.92 |
Afmid |
arylformamidase |
1003 |
0.31 |
| chr9_44387040_44387229 | 0.86 |
Hyou1 |
hypoxia up-regulated 1 |
3527 |
0.07 |
| chr3_138233542_138233745 | 0.84 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
5592 |
0.13 |
| chr5_27996032_27996183 | 0.82 |
Gm4865 |
predicted gene 4865 |
5199 |
0.19 |
| chr16_90199102_90199289 | 0.77 |
Gm49704 |
predicted gene, 49704 |
1047 |
0.47 |
| chr17_64644726_64644989 | 0.75 |
Man2a1 |
mannosidase 2, alpha 1 |
44121 |
0.17 |
| chr18_49980030_49980460 | 0.75 |
Tnfaip8 |
tumor necrosis factor, alpha-induced protein 8 |
710 |
0.72 |
| chr1_182763696_182764002 | 0.73 |
Susd4 |
sushi domain containing 4 |
11 |
0.98 |
| chr11_110177974_110178125 | 0.70 |
Abca9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
9853 |
0.25 |
| chr5_96161523_96161844 | 0.68 |
Cnot6l |
CCR4-NOT transcription complex, subunit 6-like |
59 |
0.98 |
| chr3_136670359_136670659 | 0.68 |
Ppp3ca |
protein phosphatase 3, catalytic subunit, alpha isoform |
261 |
0.94 |
| chr4_106763916_106764094 | 0.68 |
Acot11 |
acyl-CoA thioesterase 11 |
7551 |
0.15 |
| chr3_97645418_97646002 | 0.67 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
12483 |
0.13 |
| chr9_30820894_30821215 | 0.67 |
Gm31013 |
predicted gene, 31013 |
39575 |
0.16 |
| chr1_127730419_127730668 | 0.63 |
Acmsd |
amino carboxymuconate semialdehyde decarboxylase |
1130 |
0.5 |
| chr7_126273472_126273659 | 0.63 |
Sbk1 |
SH3-binding kinase 1 |
165 |
0.92 |
| chr8_117316911_117317193 | 0.58 |
Cmip |
c-Maf inducing protein |
32118 |
0.2 |
| chr13_34635371_34635522 | 0.57 |
Pxdc1 |
PX domain containing 1 |
13122 |
0.13 |
| chr11_81371218_81371402 | 0.56 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
13703 |
0.29 |
| chr19_38399310_38399568 | 0.56 |
Slc35g1 |
solute carrier family 35, member G1 |
3398 |
0.2 |
| chr9_44083899_44084207 | 0.56 |
Usp2 |
ubiquitin specific peptidase 2 |
886 |
0.32 |
| chr13_60664706_60665228 | 0.55 |
Gm48583 |
predicted gene, 48583 |
23177 |
0.15 |
| chrX_94541280_94541431 | 0.55 |
Maged1 |
melanoma antigen, family D, 1 |
635 |
0.62 |
| chr6_137797018_137797169 | 0.54 |
Dera |
deoxyribose-phosphate aldolase (putative) |
259 |
0.94 |
| chr3_31970158_31970309 | 0.53 |
Gm37834 |
predicted gene, 37834 |
15469 |
0.27 |
| chr1_155808676_155809110 | 0.53 |
Qsox1 |
quiescin Q6 sulfhydryl oxidase 1 |
3927 |
0.15 |
| chr17_57056615_57056782 | 0.53 |
Slc25a23 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
1313 |
0.19 |
| chrX_85734642_85734808 | 0.51 |
Gk |
glycerol kinase |
2885 |
0.21 |
| chr16_24517847_24518116 | 0.51 |
Morf4l1-ps1 |
mortality factor 4 like 1, pseudogene 1 |
11413 |
0.25 |
| chr1_154117784_154118141 | 0.50 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
11 |
0.98 |
| chr6_136449342_136449720 | 0.50 |
Gm25882 |
predicted gene, 25882 |
31719 |
0.1 |
| chr11_110496442_110496593 | 0.49 |
Map2k6 |
mitogen-activated protein kinase kinase 6 |
17478 |
0.25 |
| chr10_80674829_80675669 | 0.48 |
Mknk2 |
MAP kinase-interacting serine/threonine kinase 2 |
1044 |
0.31 |
| chr3_130999761_130999912 | 0.48 |
A430072C10Rik |
RIKEN cDNA A430072C10 gene |
26534 |
0.14 |
| chr5_25015695_25015994 | 0.47 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
20091 |
0.17 |
| chr8_25776385_25776540 | 0.47 |
Bag4 |
BCL2-associated athanogene 4 |
1120 |
0.32 |
| chr1_86598416_86598568 | 0.47 |
Cops7b |
COP9 signalosome subunit 7B |
1752 |
0.25 |
| chr13_9636658_9636821 | 0.47 |
Gm48888 |
predicted gene, 48888 |
350 |
0.86 |
| chr13_46735806_46736396 | 0.46 |
Nup153 |
nucleoporin 153 |
8161 |
0.17 |
| chr14_20956319_20956501 | 0.46 |
Vcl |
vinculin |
27012 |
0.18 |
| chr14_122946058_122946219 | 0.46 |
4930594M22Rik |
RIKEN cDNA 4930594M22 gene |
5981 |
0.19 |
| chr19_56533975_56534130 | 0.45 |
Dclre1a |
DNA cross-link repair 1A |
3330 |
0.24 |
| chr7_80635666_80635850 | 0.45 |
Gm15880 |
predicted gene 15880 |
259 |
0.9 |
| chr7_70798887_70799067 | 0.45 |
Gm24880 |
predicted gene, 24880 |
46561 |
0.15 |
| chr11_95385743_95385947 | 0.45 |
Slc35b1 |
solute carrier family 35, member B1 |
939 |
0.38 |
| chr7_128297307_128297458 | 0.45 |
BC017158 |
cDNA sequence BC017158 |
194 |
0.88 |
| chr8_125012381_125013106 | 0.44 |
Tsnax |
translin-associated factor X |
254 |
0.9 |
| chr16_13257357_13257584 | 0.44 |
Mrtfb |
myocardin related transcription factor B |
964 |
0.67 |
| chr1_130734318_130734676 | 0.44 |
AA986860 |
expressed sequence AA986860 |
2387 |
0.14 |
| chr9_31246201_31246667 | 0.43 |
Gm7244 |
predicted gene 7244 |
28387 |
0.13 |
| chr1_162859614_162860000 | 0.43 |
Fmo1 |
flavin containing monooxygenase 1 |
13 |
0.98 |
| chr9_106756626_106756783 | 0.43 |
Rad54l2 |
RAD54 like 2 (S. cerevisiae) |
32472 |
0.12 |
| chr13_94371154_94371305 | 0.42 |
Ap3b1 |
adaptor-related protein complex 3, beta 1 subunit |
12269 |
0.17 |
| chr18_3960517_3960692 | 0.42 |
Gm7378 |
predicted gene 7378 |
49652 |
0.15 |
| chr5_92063540_92063702 | 0.42 |
G3bp2 |
GTPase activating protein (SH3 domain) binding protein 2 |
3902 |
0.15 |
| chr13_112466453_112467116 | 0.41 |
Il6st |
interleukin 6 signal transducer |
809 |
0.59 |
| chr6_28573424_28573621 | 0.41 |
Gm37978 |
predicted gene, 37978 |
5450 |
0.18 |
| chr8_116732996_116733152 | 0.41 |
Cdyl2 |
chromodomain protein, Y chromosome-like 2 |
83 |
0.98 |
| chr8_54518329_54518486 | 0.41 |
Gm45553 |
predicted gene 45553 |
4257 |
0.2 |
| chr16_56551155_56551324 | 0.41 |
Abi3bp |
ABI gene family, member 3 (NESH) binding protein |
73339 |
0.11 |
| chr6_114089717_114089882 | 0.40 |
Gm43932 |
predicted gene, 43932 |
27603 |
0.13 |
| chr2_72222793_72223023 | 0.40 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
9394 |
0.17 |
| chr1_93433209_93433360 | 0.40 |
Hdlbp |
high density lipoprotein (HDL) binding protein |
7508 |
0.12 |
| chr14_27298945_27299301 | 0.40 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
36943 |
0.16 |
| chr17_12532229_12532396 | 0.40 |
Slc22a3 |
solute carrier family 22 (organic cation transporter), member 3 |
24608 |
0.17 |
| chr7_114088442_114088593 | 0.40 |
Gm45615 |
predicted gene 45615 |
1619 |
0.45 |
| chr8_109997497_109997880 | 0.40 |
Tat |
tyrosine aminotransferase |
7182 |
0.12 |
| chr8_46522524_46522828 | 0.40 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
4110 |
0.19 |
| chr4_135018164_135018325 | 0.39 |
Syf2 |
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
87234 |
0.06 |
| chr9_121794943_121795277 | 0.39 |
Hhatl |
hedgehog acyltransferase-like |
2603 |
0.13 |
| chr15_3512657_3512808 | 0.39 |
Ghr |
growth hormone receptor |
41088 |
0.19 |
| chr9_108297063_108297481 | 0.38 |
Amt |
aminomethyltransferase |
350 |
0.72 |
| chr8_22878603_22878765 | 0.38 |
Gm45555 |
predicted gene 45555 |
5095 |
0.18 |
| chr8_70832381_70832563 | 0.38 |
Arrdc2 |
arrestin domain containing 2 |
4115 |
0.09 |
| chr14_34251550_34251824 | 0.38 |
Gm18813 |
predicted gene, 18813 |
8645 |
0.09 |
| chr4_108782032_108782195 | 0.38 |
Zfyve9 |
zinc finger, FYVE domain containing 9 |
1315 |
0.38 |
| chr5_125323526_125323720 | 0.38 |
Gm42633 |
predicted gene 42633 |
2179 |
0.22 |
| chr1_106013874_106014051 | 0.37 |
Zcchc2 |
zinc finger, CCHC domain containing 2 |
2218 |
0.27 |
| chr18_54987200_54987598 | 0.37 |
Zfp608 |
zinc finger protein 608 |
2767 |
0.26 |
| chr9_30856213_30856364 | 0.37 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
53369 |
0.12 |
| chr7_19018198_19018349 | 0.37 |
Foxa3 |
forkhead box A3 |
5265 |
0.08 |
| chr13_55211103_55211935 | 0.37 |
Nsd1 |
nuclear receptor-binding SET-domain protein 1 |
22 |
0.97 |
| chr5_123661837_123661995 | 0.37 |
Clip1 |
CAP-GLY domain containing linker protein 1 |
4774 |
0.13 |
| chr8_84147623_84147789 | 0.36 |
Cc2d1a |
coiled-coil and C2 domain containing 1A |
159 |
0.65 |
| chr6_94163293_94163487 | 0.36 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
119635 |
0.06 |
| chr10_122380580_122380731 | 0.36 |
Gm36041 |
predicted gene, 36041 |
6237 |
0.25 |
| chr18_84054624_84054776 | 0.36 |
Tshz1 |
teashirt zinc finger family member 1 |
30375 |
0.14 |
| chr9_73983069_73983229 | 0.36 |
Unc13c |
unc-13 homolog C |
14183 |
0.25 |
| chr4_49534864_49535018 | 0.36 |
Aldob |
aldolase B, fructose-bisphosphate |
3925 |
0.16 |
| chr15_80682694_80682863 | 0.36 |
Fam83f |
family with sequence similarity 83, member F |
10931 |
0.13 |
| chr8_22934556_22934721 | 0.36 |
Kat6a |
K(lysine) acetyltransferase 6A |
20852 |
0.14 |
| chr15_59058246_59058397 | 0.35 |
Mtss1 |
MTSS I-BAR domain containing 1 |
2857 |
0.31 |
| chr1_78661192_78661343 | 0.35 |
Utp14b |
UTP14B small subunit processome component |
2827 |
0.23 |
| chr14_16380093_16380270 | 0.35 |
Top2b |
topoisomerase (DNA) II beta |
9396 |
0.19 |
| chr9_70069929_70070289 | 0.35 |
Gm47233 |
predicted gene, 47233 |
242 |
0.89 |
| chr5_52945261_52945434 | 0.35 |
Gm23532 |
predicted gene, 23532 |
14115 |
0.15 |
| chr13_29780744_29780904 | 0.35 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
74607 |
0.12 |
| chr19_32798500_32798700 | 0.35 |
Pten |
phosphatase and tensin homolog |
12625 |
0.25 |
| chr2_152733395_152733571 | 0.35 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
2768 |
0.15 |
| chr3_79219143_79219307 | 0.35 |
4921511C10Rik |
RIKEN cDNA 4921511C10 gene |
6306 |
0.2 |
| chr1_91444586_91444906 | 0.35 |
Per2 |
period circadian clock 2 |
5119 |
0.13 |
| chr1_162860588_162860757 | 0.35 |
Fmo1 |
flavin containing monooxygenase 1 |
703 |
0.67 |
| chr2_18415947_18416098 | 0.34 |
Dnajc1 |
DnaJ heat shock protein family (Hsp40) member C1 |
23192 |
0.21 |
| chr7_44246179_44246345 | 0.34 |
1700028J19Rik |
RIKEN cDNA 1700028J19 gene |
433 |
0.26 |
| chr16_91452124_91452326 | 0.34 |
Gm46562 |
predicted gene, 46562 |
6196 |
0.1 |
| chr1_105176971_105177328 | 0.34 |
Gm29012 |
predicted gene 29012 |
60695 |
0.12 |
| chr13_63231532_63231825 | 0.34 |
Aopep |
aminopeptidase O |
8351 |
0.11 |
| chr3_51378174_51378338 | 0.34 |
Gm5103 |
predicted gene 5103 |
180 |
0.9 |
| chr18_56413923_56414074 | 0.34 |
Gramd3 |
GRAM domain containing 3 |
5172 |
0.22 |
| chr2_58789663_58789814 | 0.34 |
Upp2 |
uridine phosphorylase 2 |
24413 |
0.18 |
| chr3_10022522_10022722 | 0.34 |
Gm38335 |
predicted gene, 38335 |
2917 |
0.22 |
| chr3_28709033_28709184 | 0.34 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
11091 |
0.16 |
| chr16_24438794_24438956 | 0.33 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
9216 |
0.2 |
| chr2_35385496_35385654 | 0.33 |
4930402F06Rik |
RIKEN cDNA 4930402F06 gene |
11482 |
0.13 |
| chr12_17376541_17376855 | 0.33 |
Mir3066 |
microRNA 3066 |
21306 |
0.14 |
| chr4_108060521_108060686 | 0.33 |
Scp2 |
sterol carrier protein 2, liver |
10760 |
0.13 |
| chr12_54163231_54163399 | 0.33 |
Egln3 |
egl-9 family hypoxia-inducible factor 3 |
40545 |
0.13 |
| chr10_28228240_28228413 | 0.33 |
Gm22370 |
predicted gene, 22370 |
14205 |
0.29 |
| chr5_147320087_147320238 | 0.33 |
Urad |
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
2278 |
0.17 |
| chr11_60196823_60197490 | 0.33 |
Mir6921 |
microRNA 6921 |
3461 |
0.14 |
| chr3_27127029_27127188 | 0.32 |
Ect2 |
ect2 oncogene |
4416 |
0.19 |
| chr5_43350873_43351056 | 0.32 |
Gm43020 |
predicted gene 43020 |
4295 |
0.22 |
| chr16_93932873_93933033 | 0.32 |
Cldn14 |
claudin 14 |
3136 |
0.2 |
| chr7_75619069_75619239 | 0.32 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
4466 |
0.23 |
| chr19_4148699_4149239 | 0.32 |
Coro1b |
coronin, actin binding protein 1B |
302 |
0.67 |
| chr1_72871736_72871887 | 0.32 |
Igfbp5 |
insulin-like growth factor binding protein 5 |
2464 |
0.32 |
| chr2_166155377_166155528 | 0.32 |
Sulf2 |
sulfatase 2 |
167 |
0.95 |
| chr10_127737524_127737721 | 0.32 |
Zbtb39 |
zinc finger and BTB domain containing 39 |
1916 |
0.16 |
| chr9_119154224_119154644 | 0.32 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
2659 |
0.16 |
| chr8_47673690_47674298 | 0.32 |
Ing2 |
inhibitor of growth family, member 2 |
305 |
0.8 |
| chr9_57719905_57720075 | 0.32 |
Edc3 |
enhancer of mRNA decapping 3 |
11423 |
0.12 |
| chr8_35630042_35630217 | 0.32 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
22852 |
0.17 |
| chr19_29046369_29046553 | 0.32 |
1700018L02Rik |
RIKEN cDNA 1700018L02 gene |
216 |
0.87 |
| chr19_44393747_44393961 | 0.32 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12836 |
0.14 |
| chr6_71160777_71161206 | 0.31 |
Gm44427 |
predicted gene, 44427 |
3808 |
0.16 |
| chr9_11315591_11315762 | 0.31 |
Gm18934 |
predicted gene, 18934 |
37633 |
0.2 |
| chr1_91437326_91437628 | 0.31 |
Per2 |
period circadian clock 2 |
12388 |
0.11 |
| chr11_120236241_120236693 | 0.31 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
22 |
0.83 |
| chr6_54231812_54232155 | 0.31 |
Gm15527 |
predicted gene 15527 |
22928 |
0.16 |
| chr18_60747476_60747627 | 0.31 |
Rps14 |
ribosomal protein S14 |
120 |
0.91 |
| chr11_69367509_69368033 | 0.30 |
Chd3 |
chromodomain helicase DNA binding protein 3 |
1620 |
0.17 |
| chr3_131312138_131312323 | 0.30 |
Cyp2u1 |
cytochrome P450, family 2, subfamily u, polypeptide 1 |
9003 |
0.14 |
| chr6_33306096_33306272 | 0.30 |
Exoc4 |
exocyst complex component 4 |
26171 |
0.2 |
| chr4_150242219_150242406 | 0.30 |
Eno1 |
enolase 1, alpha non-neuron |
1174 |
0.4 |
| chr6_90334778_90334975 | 0.30 |
Uroc1 |
urocanase domain containing 1 |
1587 |
0.25 |
| chr7_100077379_100077530 | 0.30 |
Pold3 |
polymerase (DNA-directed), delta 3, accessory subunit |
12583 |
0.16 |
| chr17_64720655_64720806 | 0.30 |
Man2a1 |
mannosidase 2, alpha 1 |
7111 |
0.22 |
| chr2_85048155_85048335 | 0.30 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
190 |
0.9 |
| chr4_141134526_141134677 | 0.30 |
Szrd1 |
SUZ RNA binding domain containing 1 |
5126 |
0.12 |
| chr19_26744877_26745028 | 0.30 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
257 |
0.94 |
| chr18_53810810_53810966 | 0.30 |
Csnk1g3 |
casein kinase 1, gamma 3 |
51234 |
0.15 |
| chr7_26315480_26315657 | 0.30 |
Gm42375 |
predicted gene, 42375 |
8282 |
0.14 |
| chr3_28705261_28705416 | 0.29 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
7321 |
0.18 |
| chr4_139386772_139386990 | 0.29 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
6212 |
0.12 |
| chr14_63183364_63183533 | 0.29 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
3870 |
0.17 |
| chr9_120009962_120010210 | 0.29 |
Gm49427 |
predicted gene, 49427 |
281 |
0.82 |
| chr11_100610223_100610688 | 0.29 |
Dnajc7 |
DnaJ heat shock protein family (Hsp40) member C7 |
2902 |
0.14 |
| chr1_65174300_65174494 | 0.29 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
1167 |
0.4 |
| chr11_108308165_108308332 | 0.29 |
Apoh |
apolipoprotein H |
35106 |
0.15 |
| chr4_43656130_43656321 | 0.29 |
Hint2 |
histidine triad nucleotide binding protein 2 |
218 |
0.81 |
| chr3_79809306_79809467 | 0.29 |
Gm26420 |
predicted gene, 26420 |
25547 |
0.15 |
| chr11_4229963_4230122 | 0.29 |
Gm11956 |
predicted gene 11956 |
1223 |
0.29 |
| chr5_145139980_145140681 | 0.29 |
Bud31 |
BUD31 homolog |
32 |
0.55 |
| chr14_20174882_20175033 | 0.28 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
6852 |
0.14 |
| chr2_157130956_157131107 | 0.28 |
Samhd1 |
SAM domain and HD domain, 1 |
974 |
0.5 |
| chr9_74848568_74848768 | 0.28 |
Gm16551 |
predicted gene 16551 |
142 |
0.58 |
| chr5_8894734_8894942 | 0.28 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
889 |
0.53 |
| chr1_190117320_190117686 | 0.28 |
Gm28172 |
predicted gene 28172 |
51167 |
0.13 |
| chr7_79659672_79659823 | 0.28 |
Ticrr |
TOPBP1-interacting checkpoint and replication regulator |
449 |
0.73 |
| chr4_131883686_131883871 | 0.28 |
Srsf4 |
serine and arginine-rich splicing factor 4 |
517 |
0.65 |
| chr9_21250184_21250335 | 0.28 |
S1pr5 |
sphingosine-1-phosphate receptor 5 |
1816 |
0.16 |
| chr13_119389151_119389302 | 0.28 |
Nnt |
nicotinamide nucleotide transhydrogenase |
19726 |
0.17 |
| chrX_10719688_10719839 | 0.28 |
Gm14493 |
predicted gene 14493 |
1053 |
0.45 |
| chr4_117830286_117830539 | 0.28 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
4094 |
0.14 |
| chr18_32557156_32557525 | 0.28 |
Gypc |
glycophorin C |
2640 |
0.3 |
| chr13_12457474_12457661 | 0.27 |
Lgals8 |
lectin, galactose binding, soluble 8 |
1044 |
0.46 |
| chr3_121793319_121793679 | 0.27 |
4633401B06Rik |
RIKEN cDNA 4633401B06 gene |
4927 |
0.14 |
| chr7_141919332_141919505 | 0.27 |
Tollip |
toll interacting protein |
911 |
0.5 |
| chr2_118548783_118549158 | 0.27 |
Bmf |
BCL2 modifying factor |
692 |
0.66 |
| chr5_139775420_139775673 | 0.27 |
Ints1 |
integrator complex subunit 1 |
92 |
0.95 |
| chr10_37138598_37138749 | 0.27 |
Marcks |
myristoylated alanine rich protein kinase C substrate |
247 |
0.81 |
| chr6_85810403_85810557 | 0.27 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
617 |
0.53 |
| chr10_67058626_67058777 | 0.27 |
Reep3 |
receptor accessory protein 3 |
21379 |
0.17 |
| chr1_88042666_88042990 | 0.27 |
AC087801.1 |
UDP glycosyltransferase 1 family (Ytg1) pseudogene |
5228 |
0.08 |
| chr1_138964147_138964506 | 0.27 |
Dennd1b |
DENN/MADD domain containing 1B |
457 |
0.64 |
| chr11_95438968_95439128 | 0.27 |
Gm11522 |
predicted gene 11522 |
23673 |
0.12 |
| chr15_81394865_81395034 | 0.27 |
St13 |
suppression of tumorigenicity 13 |
346 |
0.81 |
| chr10_40300520_40300675 | 0.27 |
Amd1 |
S-adenosylmethionine decarboxylase 1 |
979 |
0.41 |
| chr2_26902336_26902522 | 0.27 |
Surf6 |
surfeit gene 6 |
361 |
0.69 |
| chr14_47512089_47512243 | 0.26 |
Gm35166 |
predicted gene, 35166 |
3979 |
0.14 |
| chr1_162871486_162871643 | 0.26 |
Fmo1 |
flavin containing monooxygenase 1 |
4954 |
0.19 |
| chr10_61138290_61138447 | 0.26 |
Sgpl1 |
sphingosine phosphate lyase 1 |
8530 |
0.15 |
| chr13_98716100_98716291 | 0.26 |
Gm9465 |
predicted gene 9465 |
315 |
0.84 |
| chr11_76997868_76998019 | 0.26 |
Slc6a4 |
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
660 |
0.62 |
| chr4_103393218_103393514 | 0.26 |
Gm12718 |
predicted gene 12718 |
10845 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.1 | 0.5 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.1 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.2 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 | 0.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.1 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.1 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.2 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.2 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.2 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.3 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.0 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.2 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.0 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.0 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:1990144 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.0 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.5 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.0 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.0 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.0 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 0.0 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.1 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) |
| 0.0 | 0.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.1 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.0 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.0 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |