Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Sample name: keratinocyteH3K4me3
QC summary table
QC statistics for this sample |
Comparison with ChIP-Seq ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP replicates: |
0.786 | 59th percentile | |
Fraction of mapped reads for the background replicates: |
0.330 | 1st percentile | |
ChIP enrichment signal intensity |
|||
Noise level ChIP signal intensity: |
0.260 | 74th percentile | |
Error in fit of enrichment distribution: |
0.350 | 9th percentile | |
Peak statistics |
|||
Number of peaks: |
116616 | 99th percentile | |
Regulatory motifs sorted by significance (z-value) for sample keratinocyteH3K4me3.
Regulatory motifs sorted by significance (z-value).
| WM name | Z-value | Associated genes | Logo |
|---|---|---|---|
| TP63 | 15.816 |
|
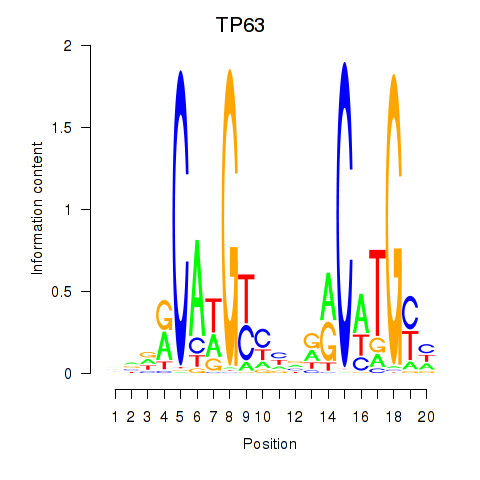
|
| ZNF711_TFAP2A_TFAP2D | 7.088 |
|
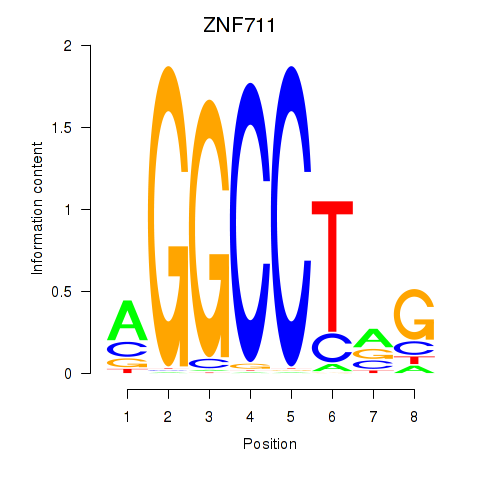
|
| ZEB1 | 6.013 |
|
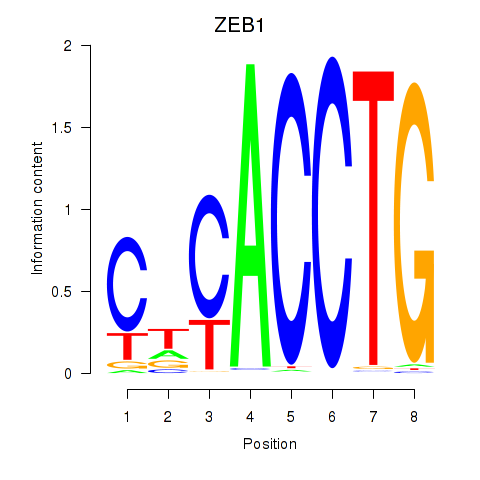
|
| SP4_PML | 5.348 |
|
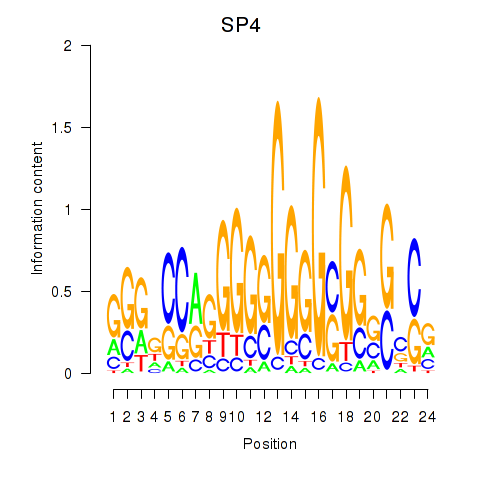
|
| PLAGL1 | 5.030 |
|
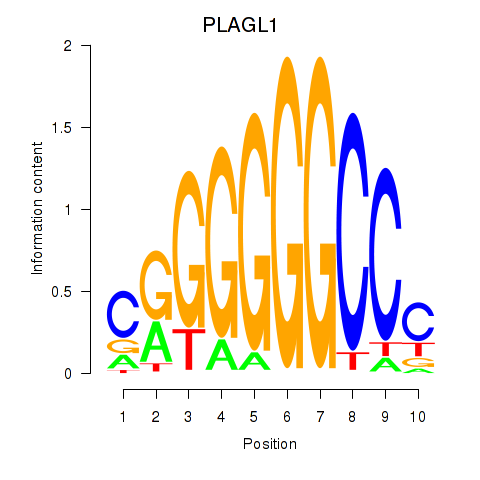
|
| RCOR1_MTA3 | 4.967 |
|
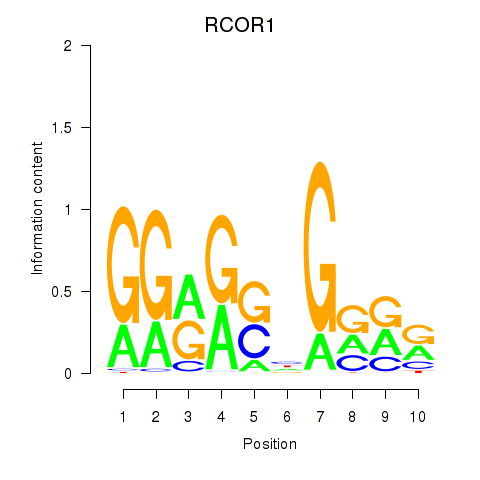
|
| FOXL1 | 4.697 |
|
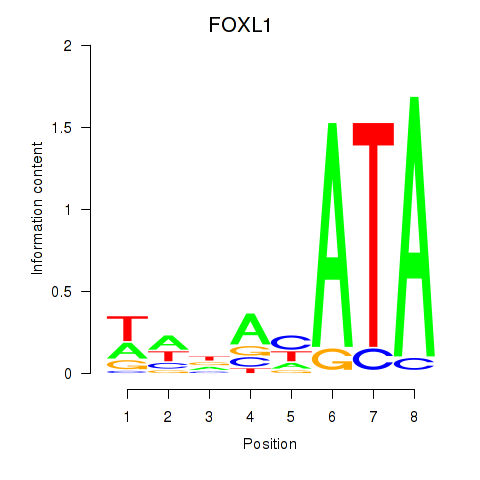
|
| TEAD3_TEAD1 | 4.332 |
|
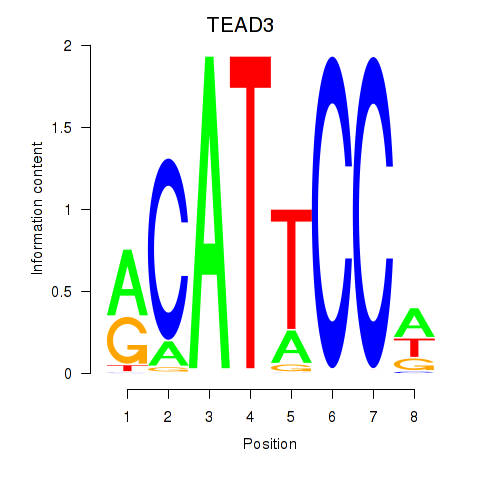
|
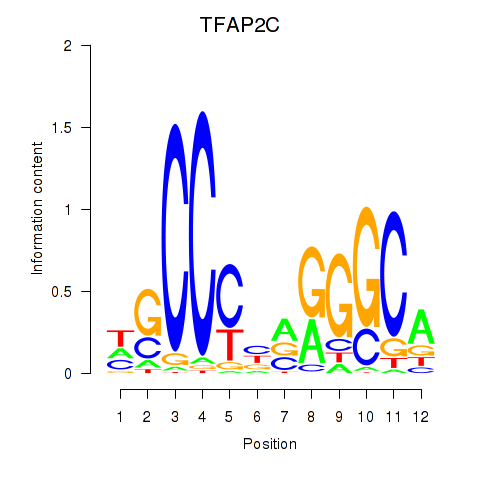
| TFAP2C | 4.302 |
|
|
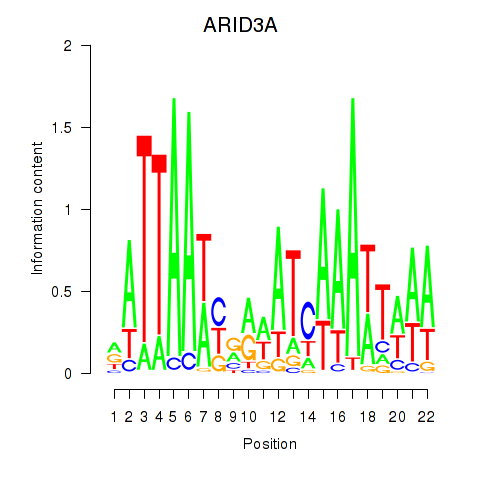
| ARID3A | 4.232 |
|
|
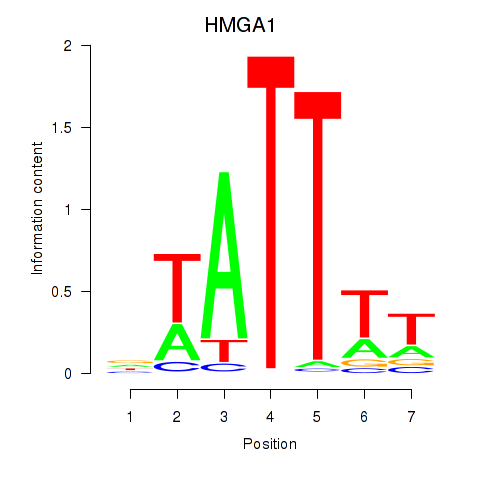
| HMGA1 | 3.980 |
|
|
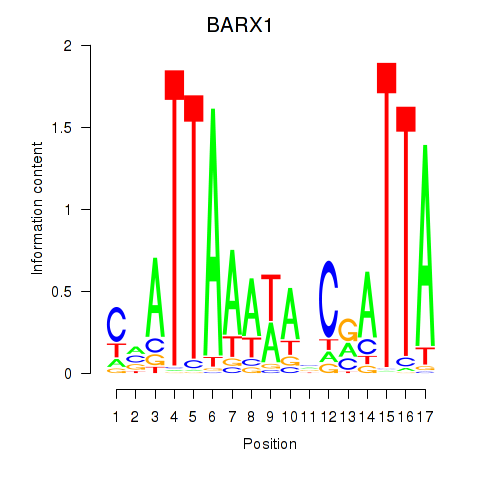
| BARX1 | 3.829 |
|
|
| TP53 | 3.676 |
|
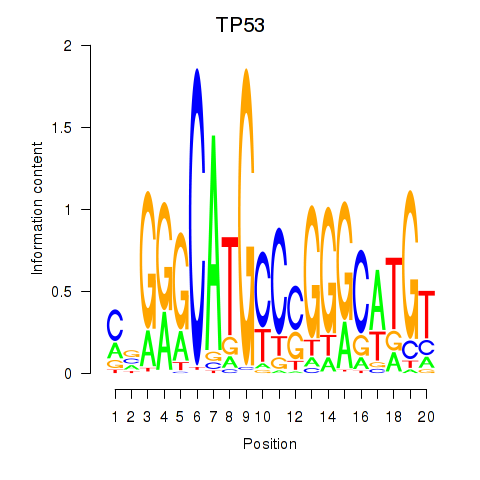
|
| ARID5A | 3.640 |
|
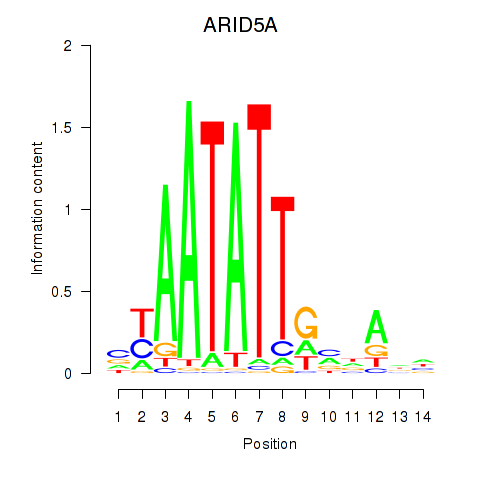
|
| PATZ1_KLF4 | 3.615 |
|
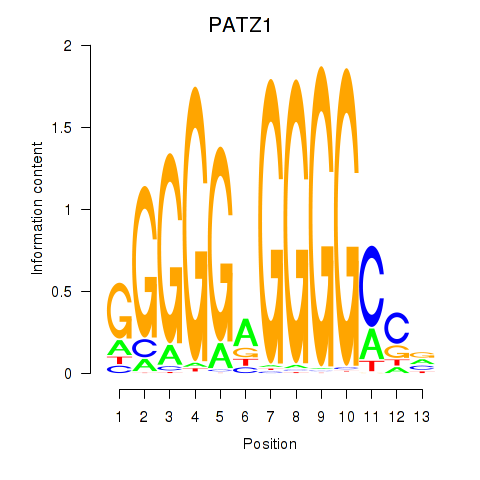
|
| WT1_MTF1_ZBTB7B | 3.594 |
|
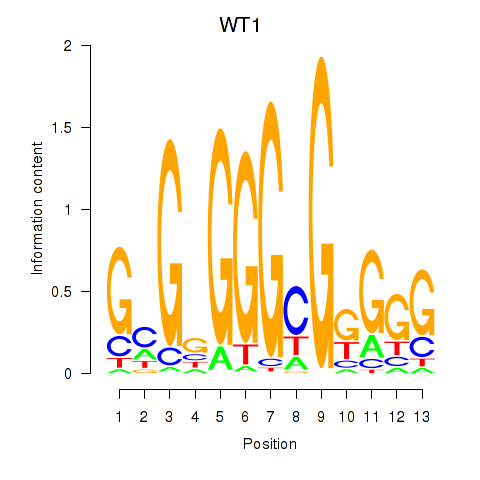
|
| CDC5L | 3.437 |
|
|
| TWIST1_SNAI1 | 3.429 |
|
|
| CUX2 | 3.366 |
|
|
| OLIG2_NEUROD1_ATOH1 | 3.318 |
|
|
| HIC2 | 2.924 |
|
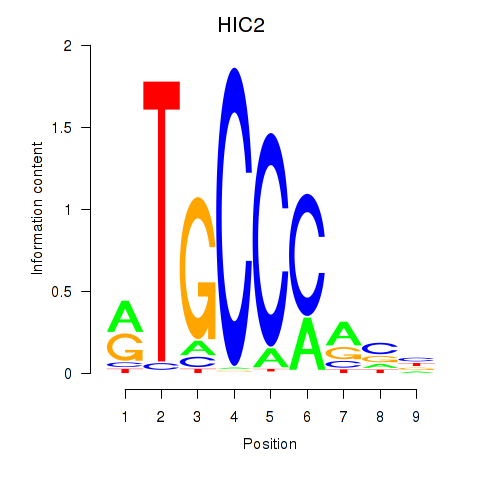
|
| NFIX_NFIB | 2.845 |
|
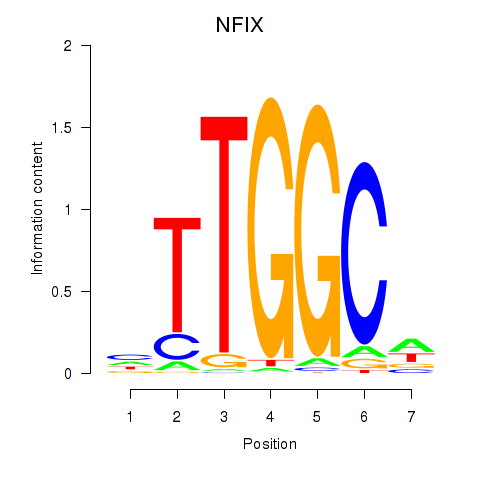
|
| SOX13_SOX12 | 2.608 |
|
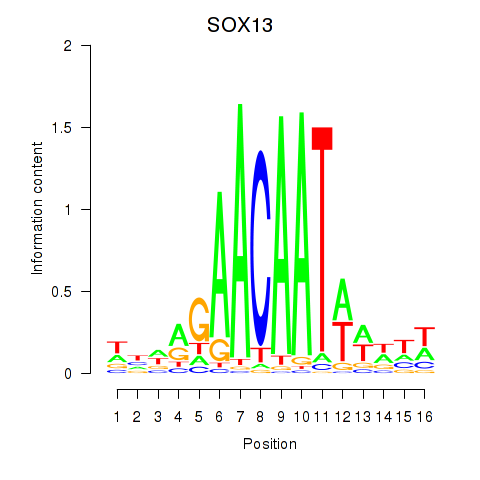
|
| GATA5 | 2.608 |
|
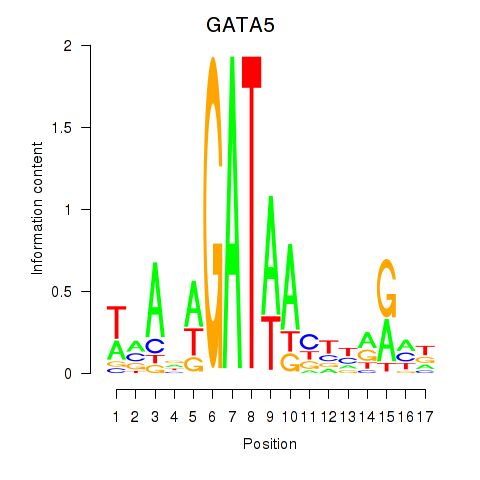
|
| SP3 | 2.521 |
|
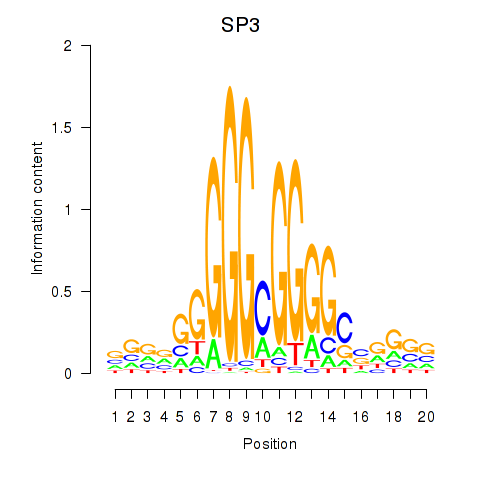
|
| MEIS3_TGIF2LX | 2.431 |
|
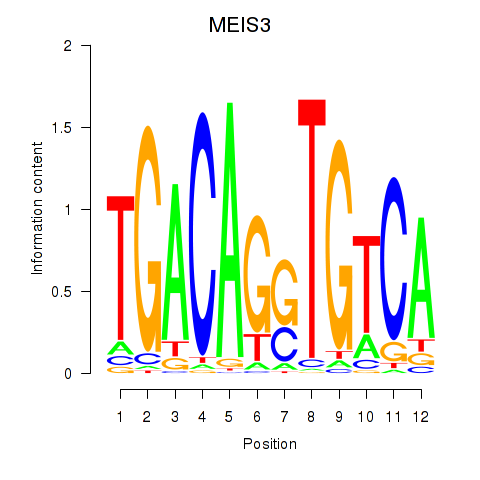
|
| E2F3 | 2.405 |
|
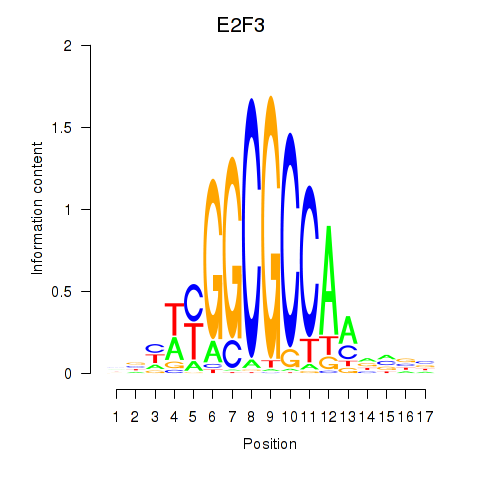
|
| TFAP2B | 2.307 |
|
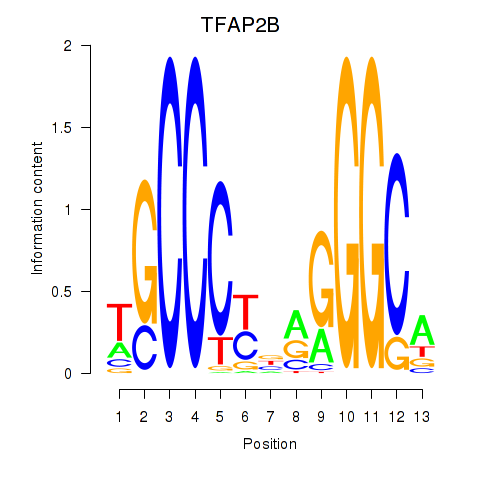
|
| POU3F3_POU3F4 | 2.230 |
|
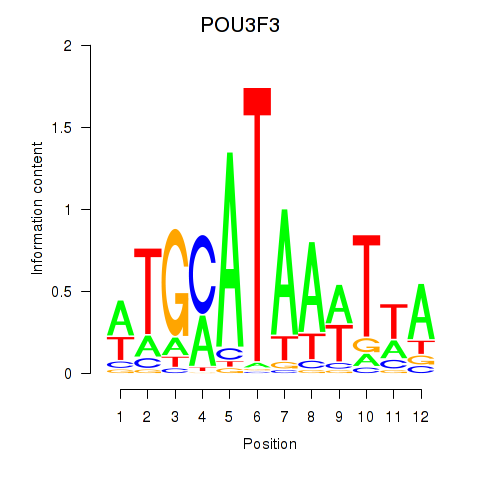
|
| KLF3 | 2.199 |
|
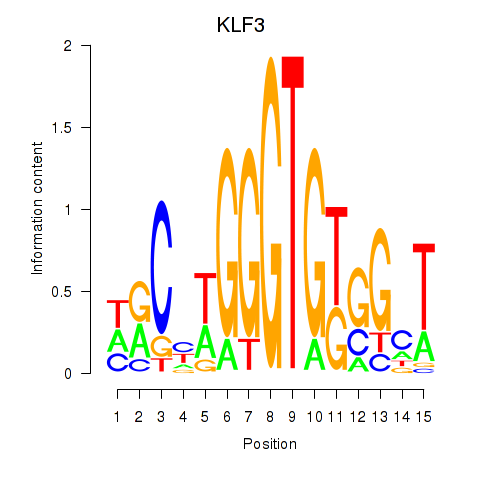
|
| FOXA2_FOXJ3 | 2.194 |
|
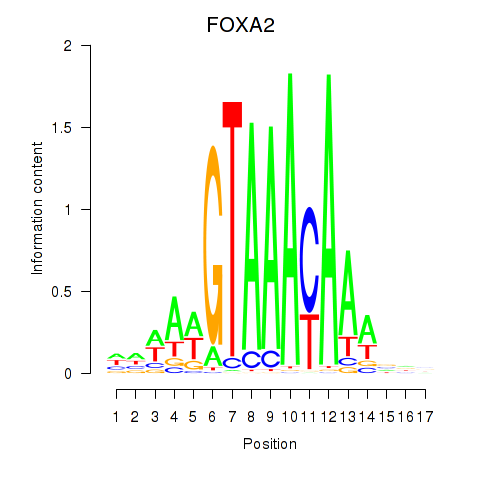
|
| TLX1_NFIC | 2.144 |
|
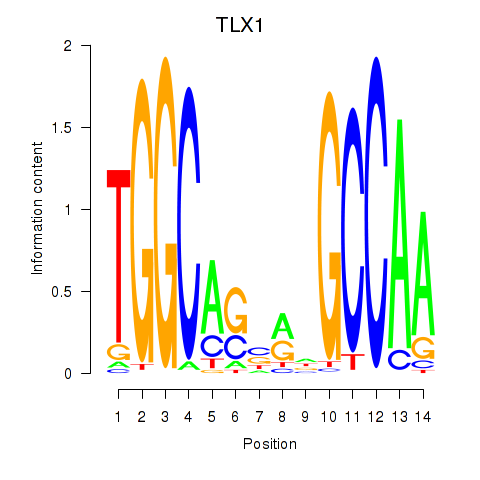
|
| SMAD4 | 2.131 |
|
|
| ZBTB16 | 2.106 |
|
|
| ZNF148 | 2.084 |
|
|
| NR2F2 | 2.063 |
|
|
| MECP2 | 2.056 |
|
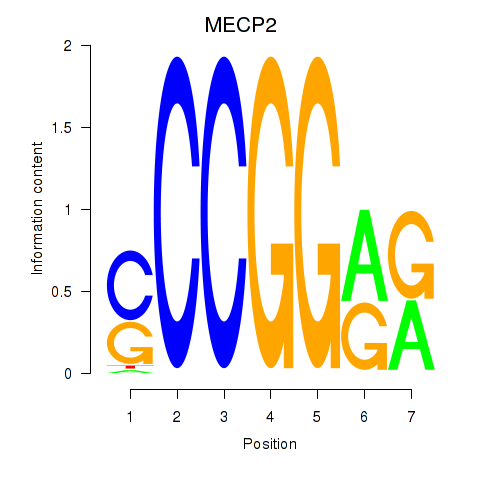
|
| ZNF143 | 1.999 |
|
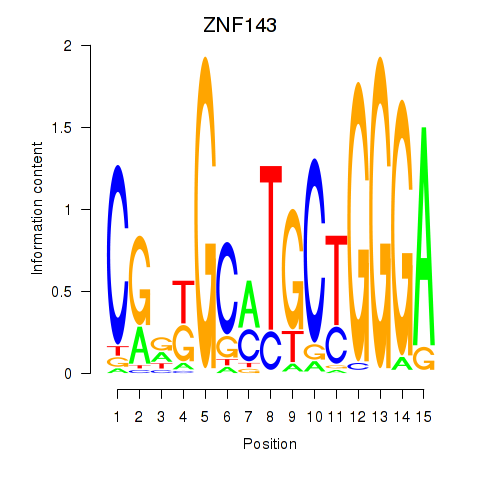
|
| GRHL1 | 1.967 |
|
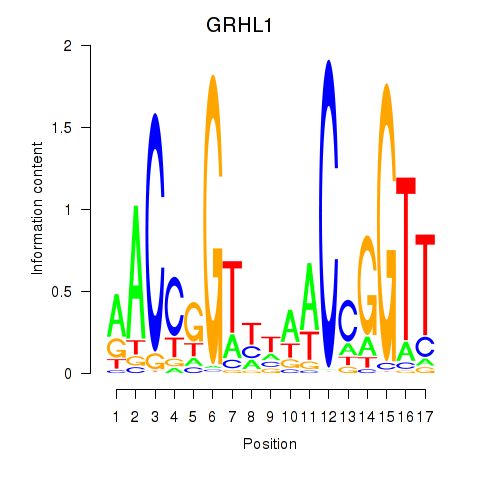
|
| KLF1 | 1.926 |
|
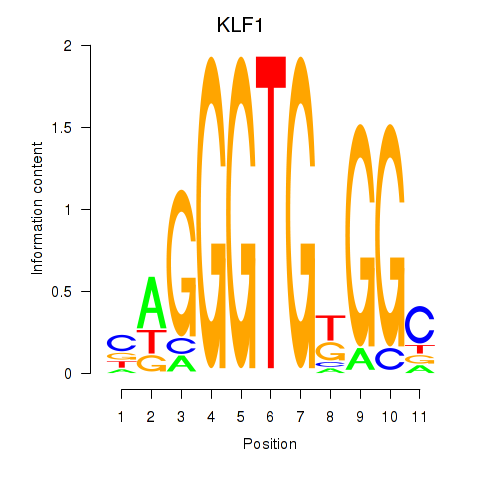
|
| ZBTB14 | 1.919 |
|
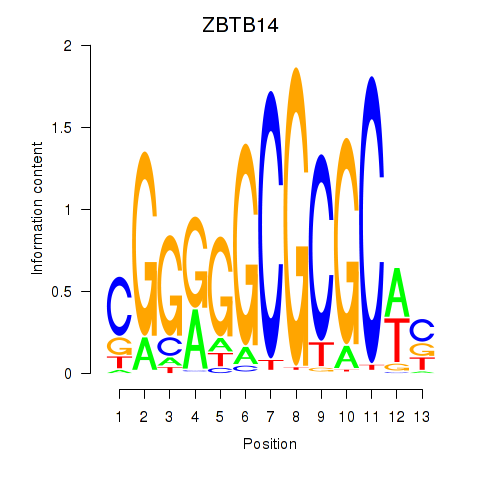
|
| HOXC10_HOXD13 | 1.859 |
|
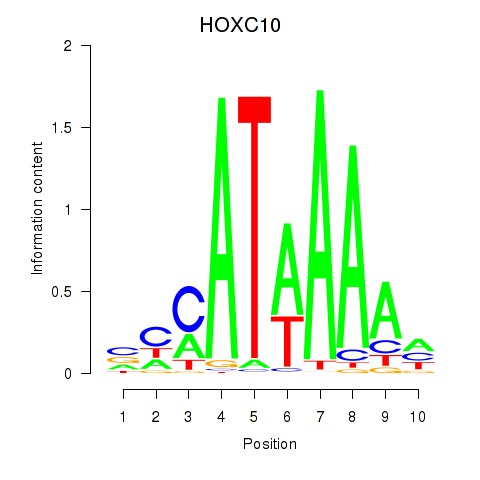
|
| JDP2 | 1.844 |
|
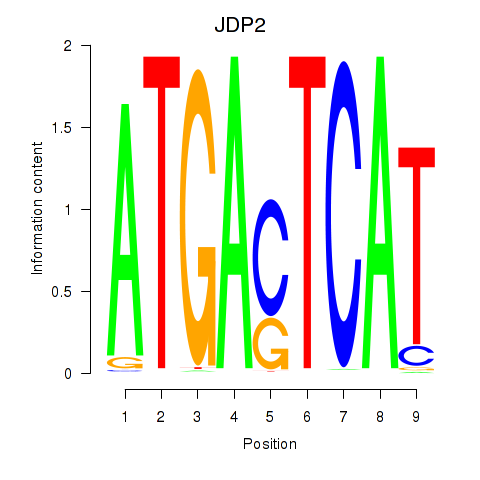
|
| ESR1 | 1.806 |
|
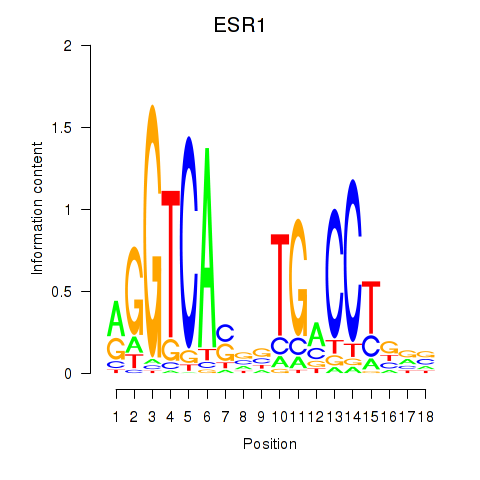
|
| GMEB1 | 1.793 |
|
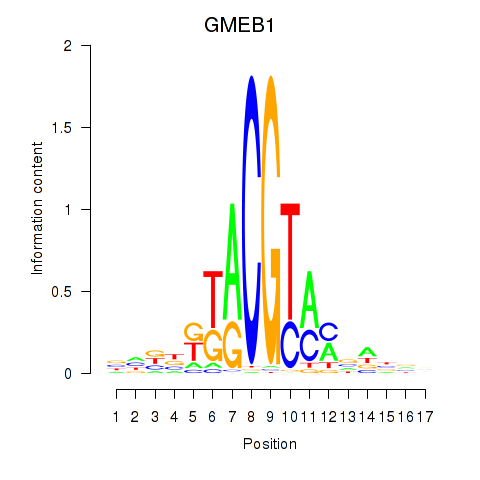
|
| MNT_HEY1_HEY2 | 1.758 |
|
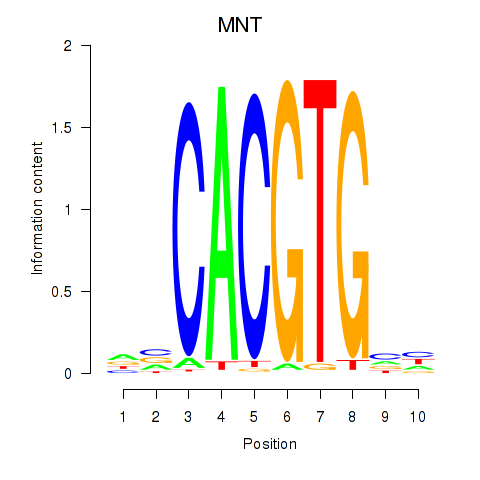
|
| FOSL2_SMARCC1 | 1.662 |
|
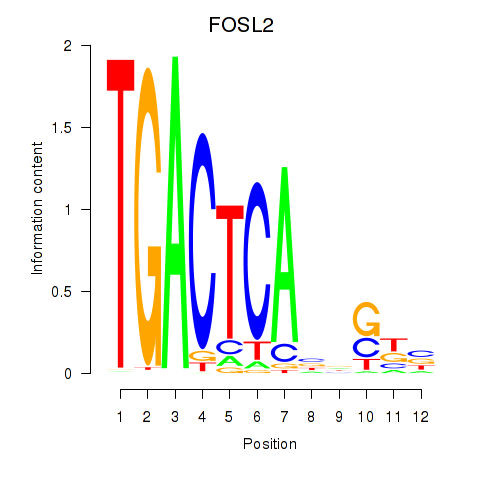
|
| HOXA9 | 1.624 |
|
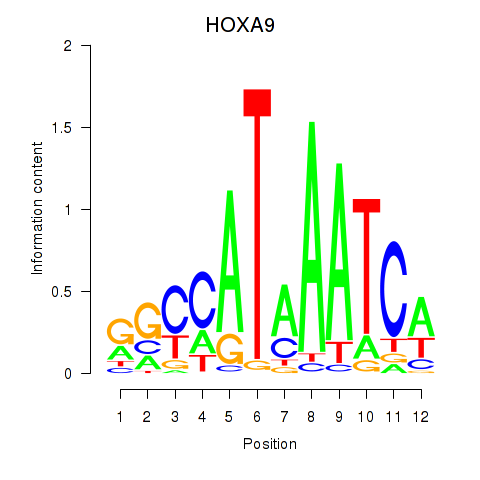
|
| MEIS1 | 1.605 |
|
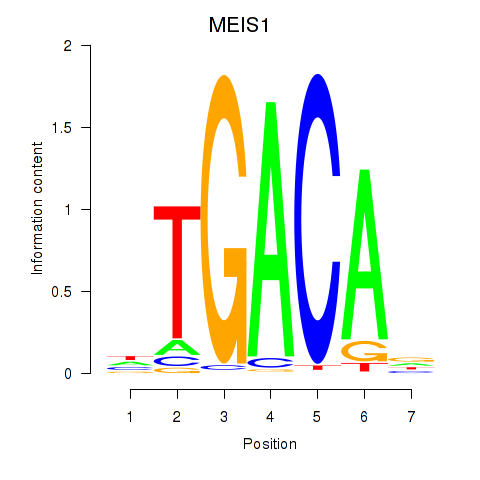
|
| MAZ_ZNF281_GTF2F1 | 1.570 |
|
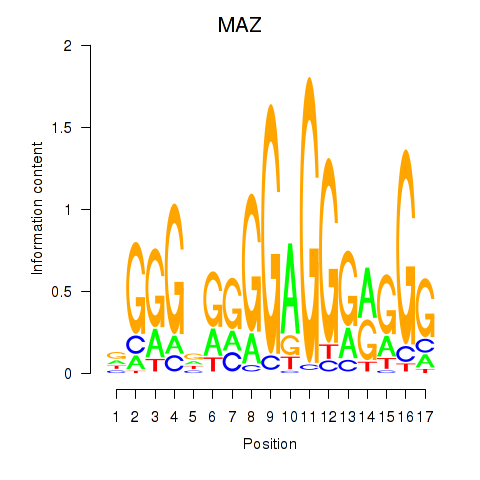
|
| POU1F1 | 1.543 |
|
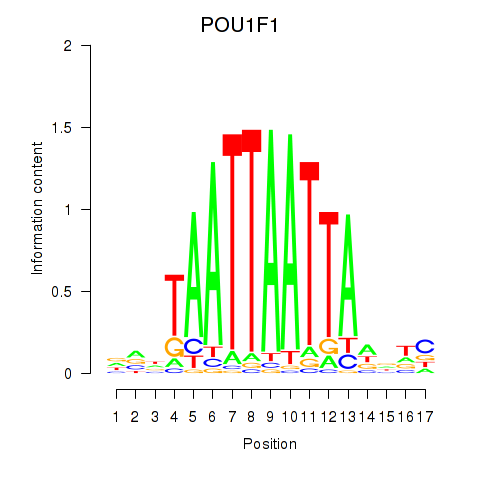
|
| DLX4_HOXD8 | 1.540 |
|
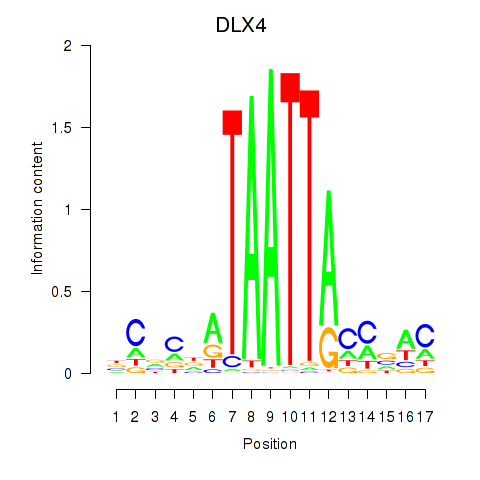
|
| IRX5 | 1.489 |
|
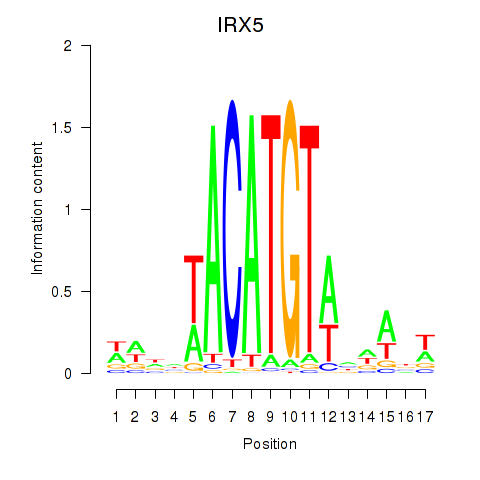
|
| PAX7_NOBOX | 1.480 |
|
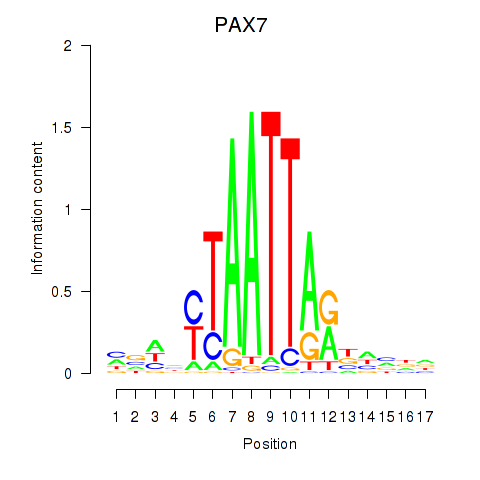
|
| MECOM | 1.467 |
|
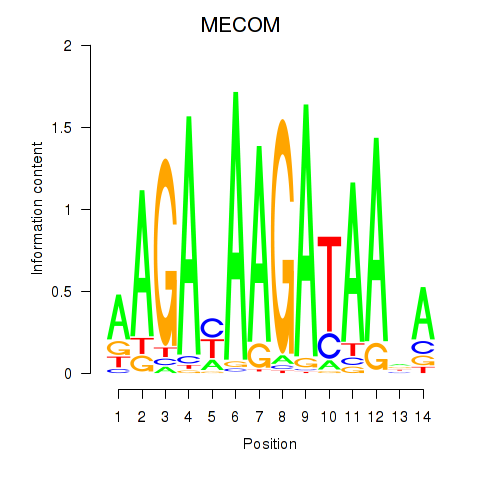
|
| HNF1A_HNF1B | 1.452 |
|
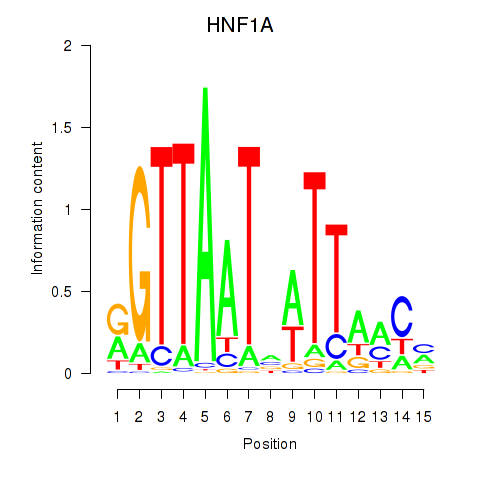
|
| POU4F2 | 1.444 |
|
|
| DBX2_HLX | 1.435 |
|
|
| EP300 | 1.420 |
|
|
| ESRRA_ESR2 | 1.413 |
|
|
| IKZF2 | 1.407 |
|
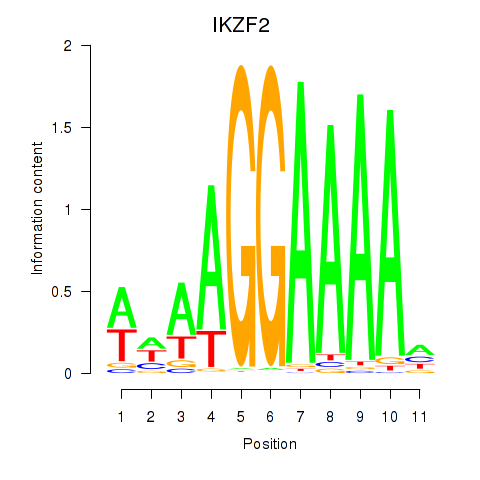
|
| HOXC13 | 1.371 |
|
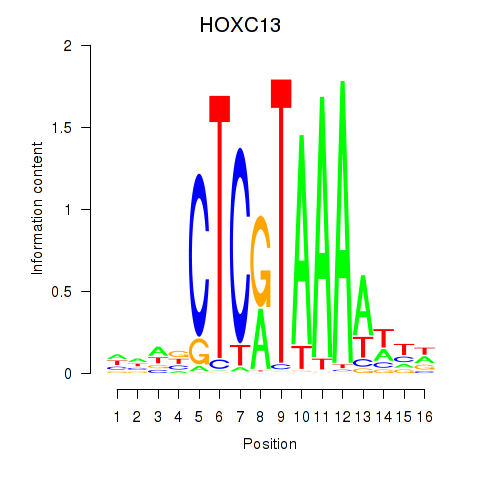
|
| TFCP2L1 | 1.362 |
|
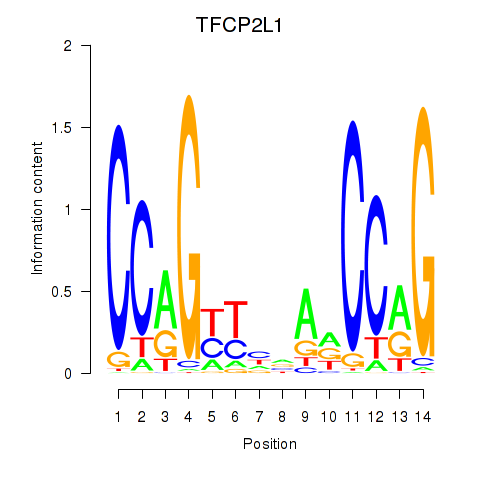
|
| LHX3 | 1.345 |
|
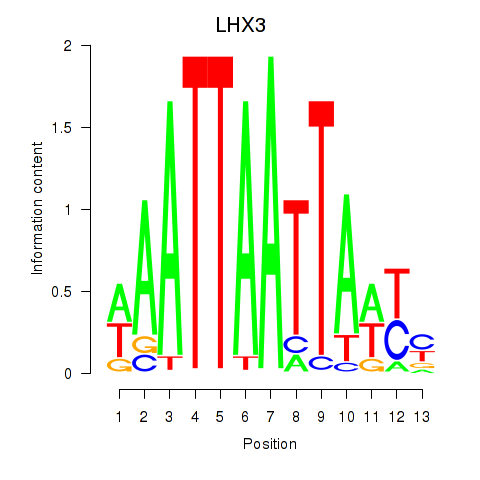
|
| EMX2 | 1.345 |
|
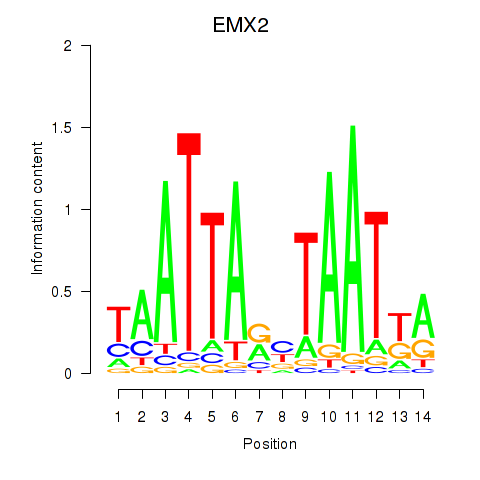
|
| PKNOX1_TGIF2 | 1.280 |
|
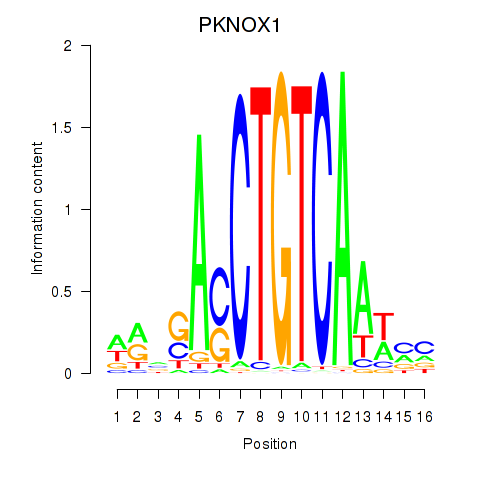
|
| ETV1_ERF_FEV_ELF1 | 1.254 |
|
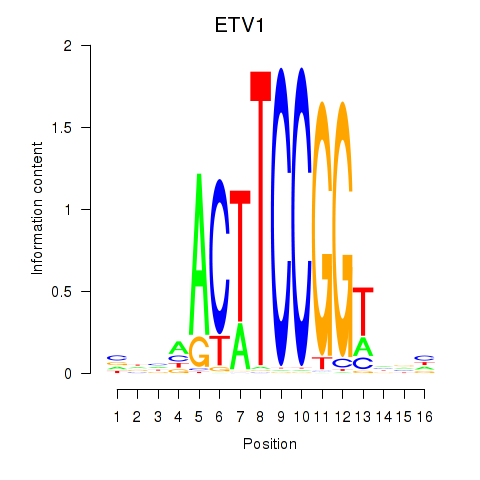
|
| NR2E1 | 1.241 |
|
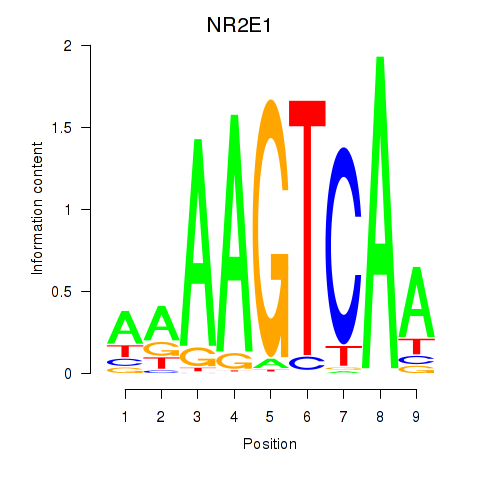
|
| POU2F1 | 1.233 |
|
|
| CEBPG | 1.230 |
|
|
| TCF12_ASCL2 | 1.217 |
|
|
| E2F4 | 1.213 |
|
|
| KLF8 | 1.210 |
|
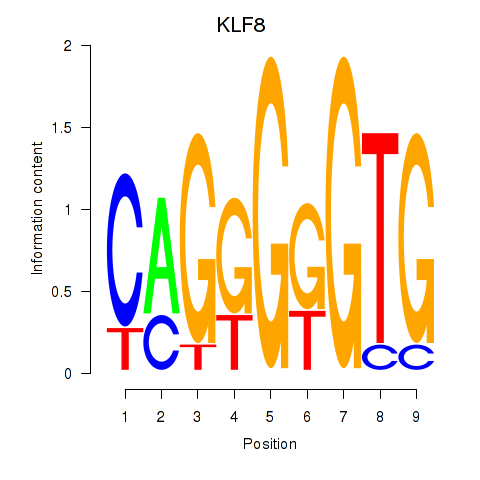
|
| ATF7 | 1.163 |
|
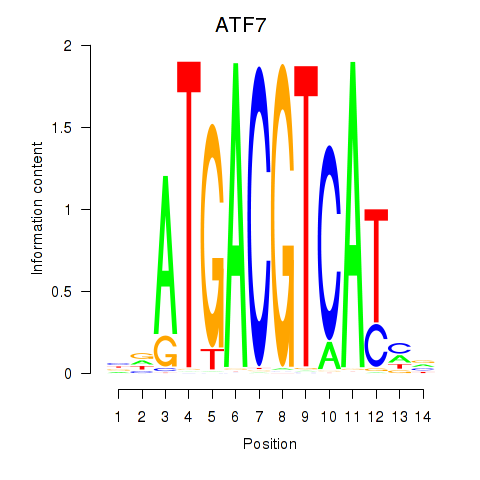
|
| NHLH1 | 1.151 |
|
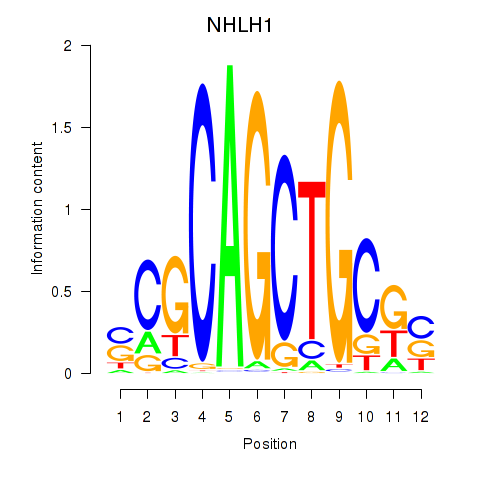
|
| MIXL1_GSX1_BSX_MEOX2_LHX4 | 1.134 |
|
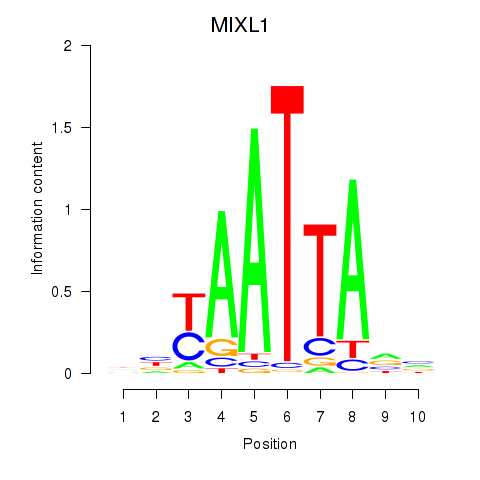
|
| GLI3 | 1.126 |
|
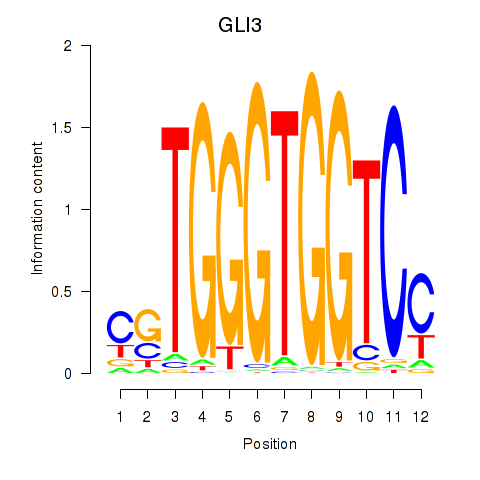
|
| TBX5 | 1.100 |
|
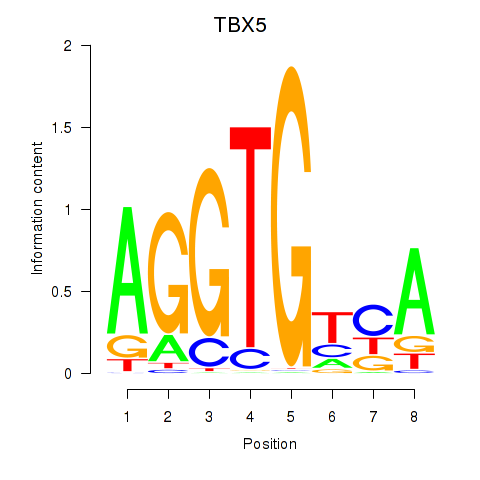
|
| HBP1 | 1.093 |
|
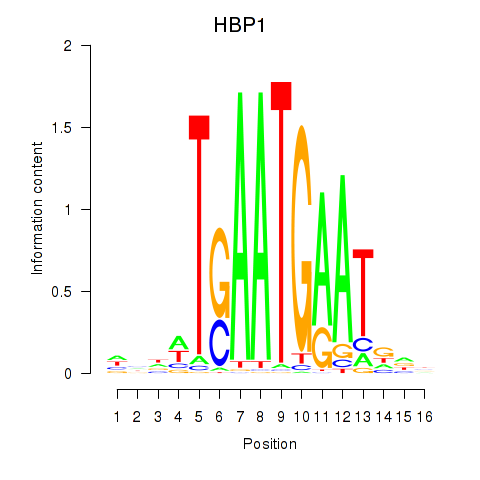
|
| ZIC1 | 1.078 |
|
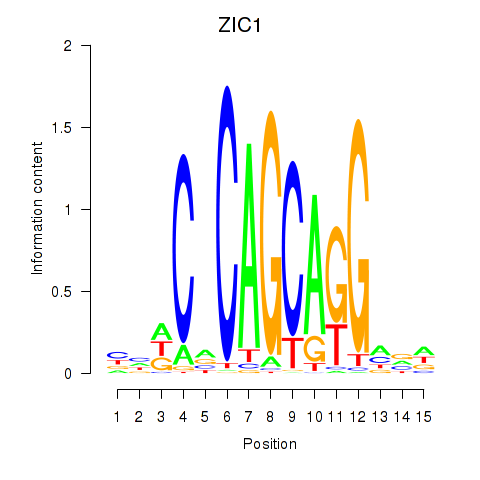
|
| MSX1 | 1.077 |
|
|
| ISL2 | 1.077 |
|
|
| TFAP4_MSC | 1.068 |
|
|
| NR1H2 | 1.064 |
|
|
| ZBTB7A_ZBTB7C | 1.056 |
|
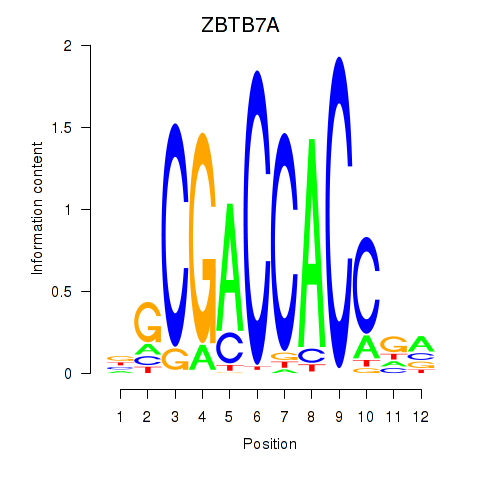
|
| MAFB | 1.051 |
|
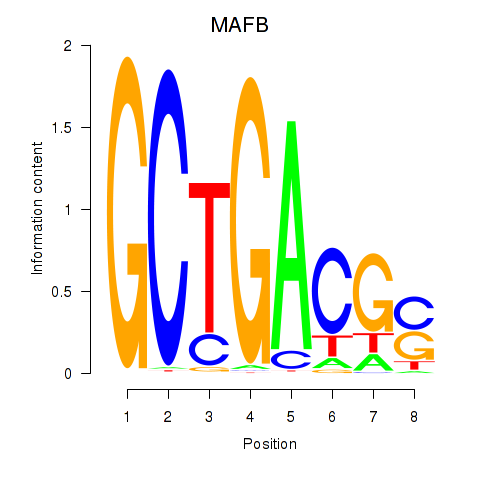
|
| GTF2I | 1.049 |
|
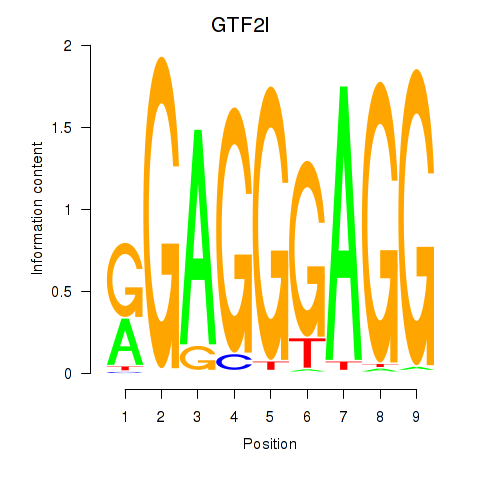
|
| HIC1 | 1.036 |
|
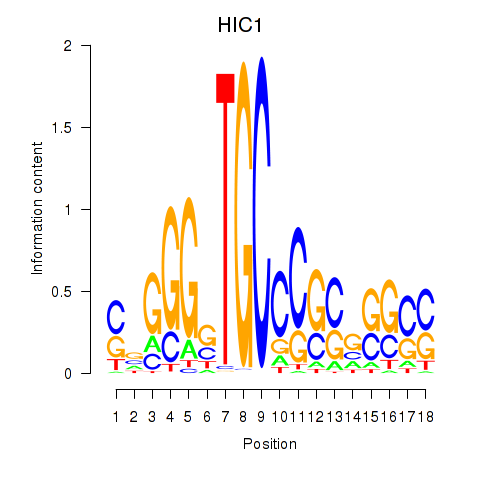
|
| CREB5_CREM_JUNB | 0.997 |
|
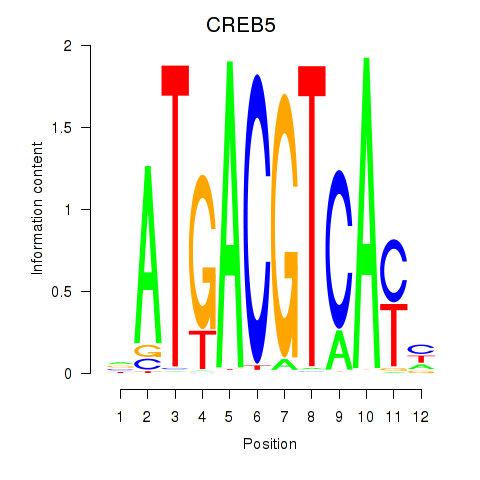
|
| TCF7L1 | 0.993 |
|
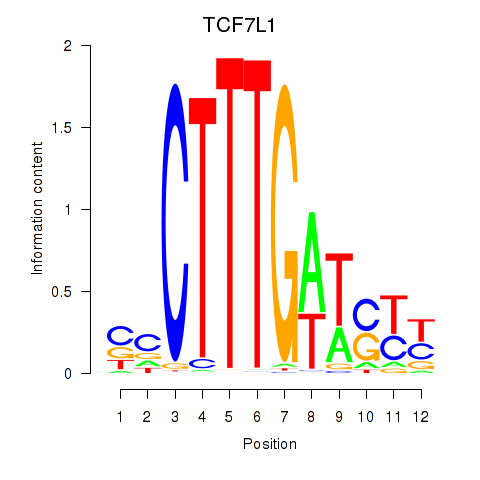
|
| HMGA2 | 0.979 |
|
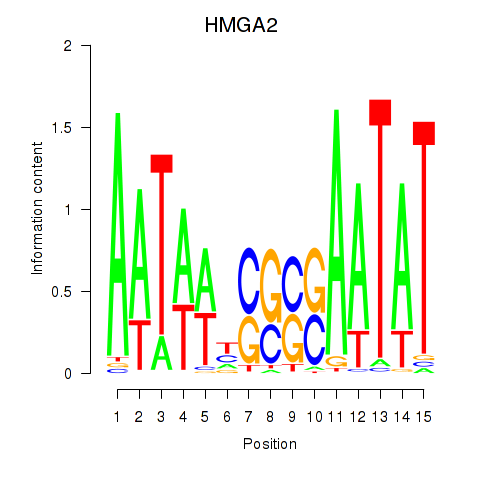
|
| EOMES | 0.948 |
|
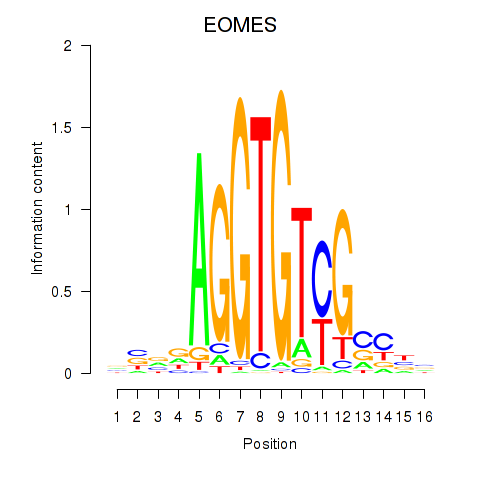
|
| IRX6_IRX4 | 0.932 |
|
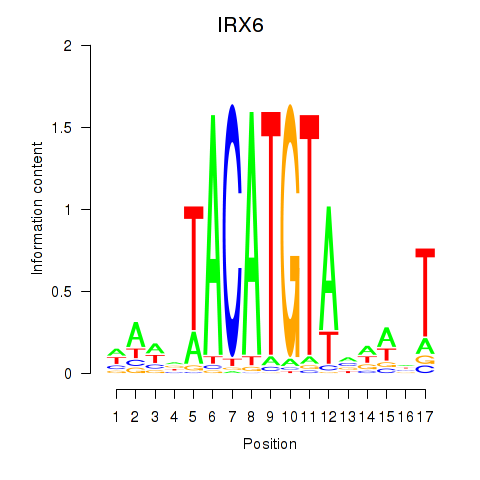
|
| HIF1A | 0.926 |
|
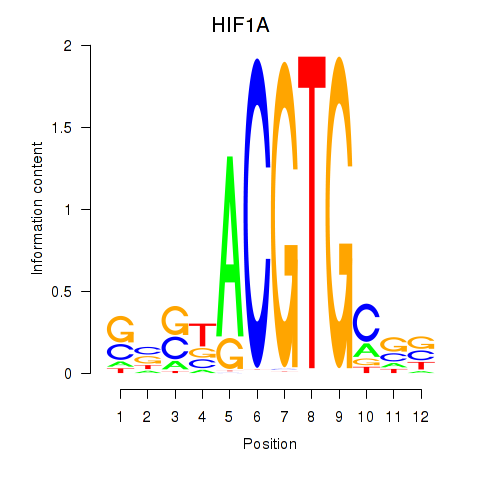
|
| HOXB3 | 0.878 |
|
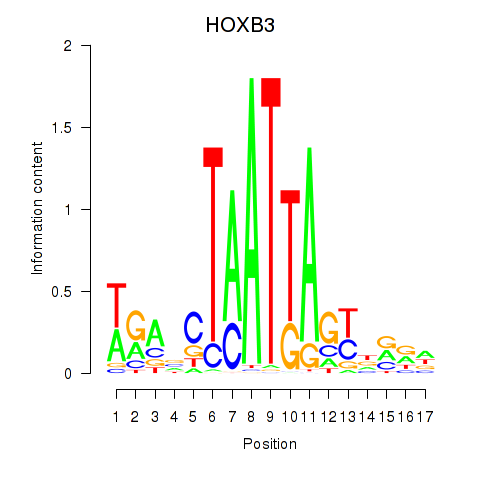
|
| OTX1 | 0.867 |
|
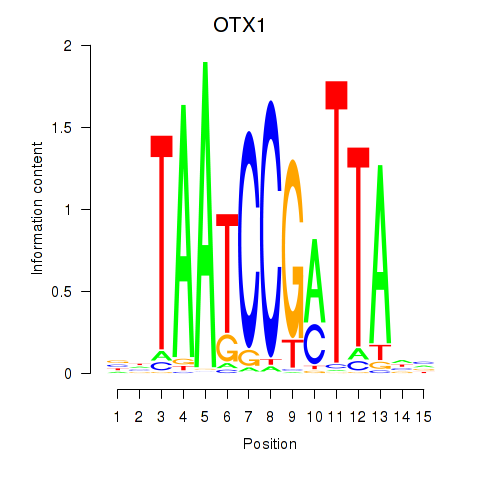
|
| RUNX3_BCL11A | 0.863 |
|
|
| MSX2 | 0.846 |
|
|
| NFKB2 | 0.831 |
|
|
| NKX2-6 | 0.815 |
|
|
| JUND | 0.805 |
|
|
| ELF3_EHF | 0.801 |
|
|
| HOXD10 | 0.794 |
|
|
| FOXQ1 | 0.777 |
|
|
| ETV6 | 0.770 |
|
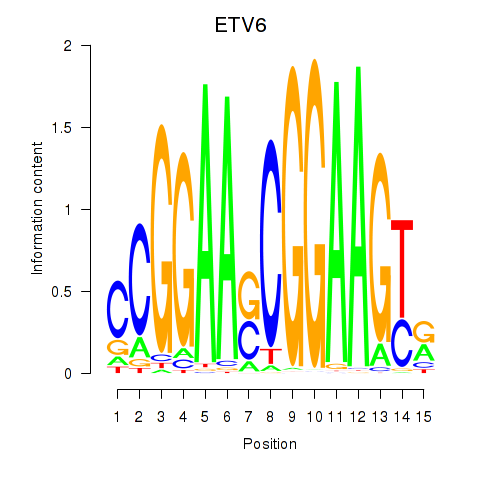
|
| REST | 0.765 |
|
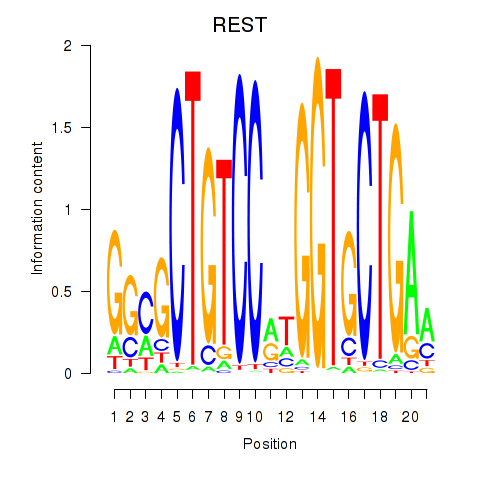
|
| HESX1 | 0.733 |
|
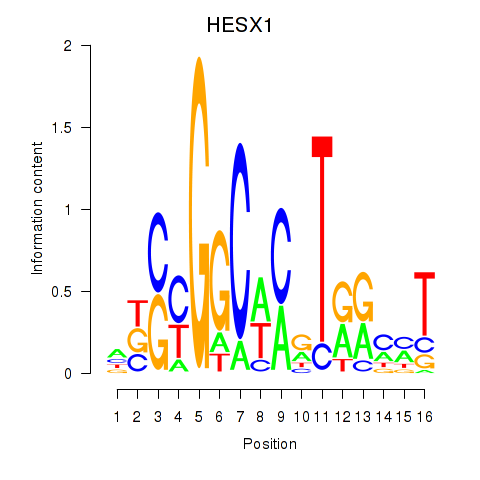
|
| NFKB1 | 0.730 |
|
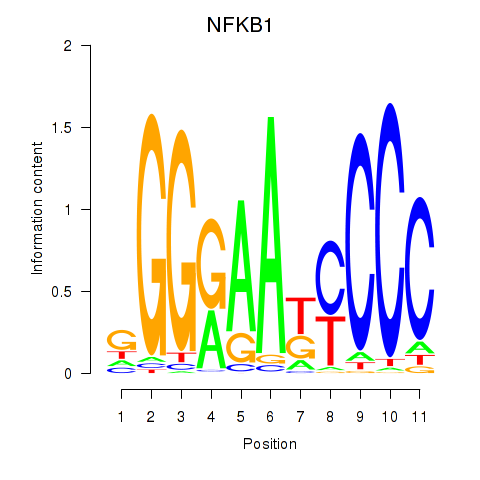
|
| NR2F1 | 0.723 |
|
|
| DUXA | 0.721 |
|
|
| AHR_ARNT2 | 0.713 |
|
|
| HOXA2_HOXB1 | 0.706 |
|
|
| SIN3A_CHD1 | 0.704 |
|
|
| HSF1 | 0.703 |
|
|
| SOX7 | 0.694 |
|
|
| SOX4 | 0.685 |
|
|
| TP73 | 0.681 |
|
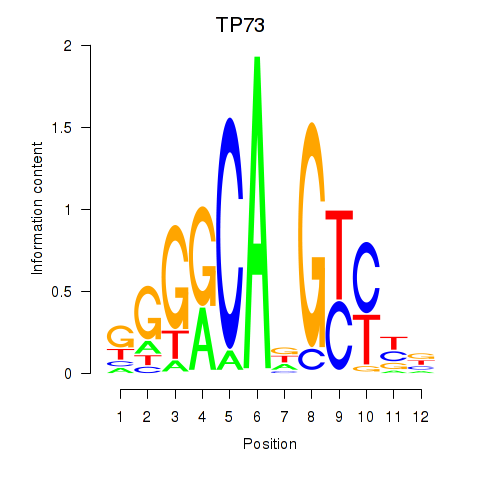
|
| VENTX | 0.678 |
|
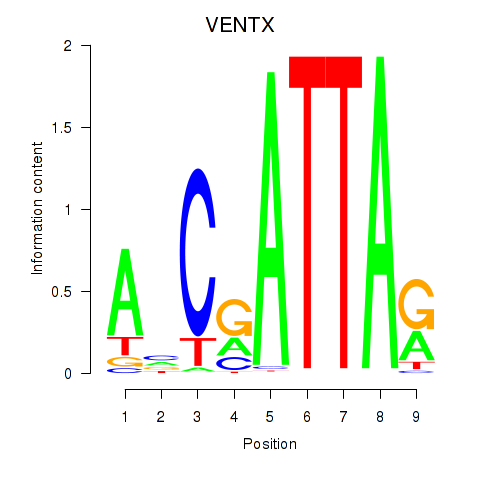
|
| DRGX_PROP1 | 0.677 |
|
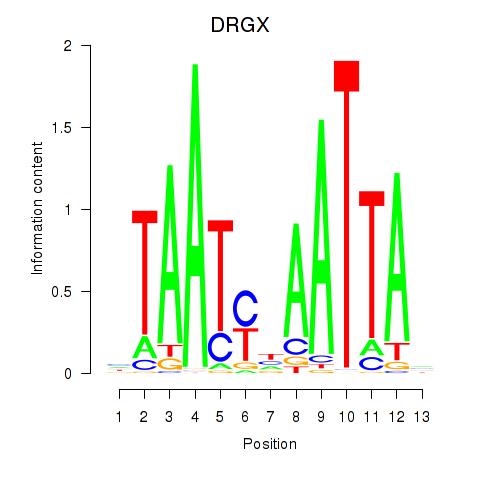
|
| SHOX | 0.672 |
|
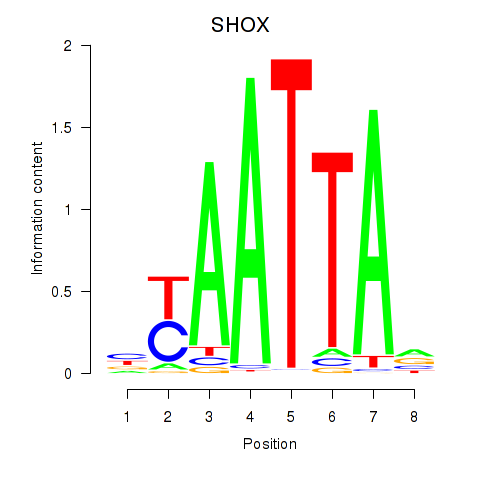
|
| SMAD2 | 0.671 |
|
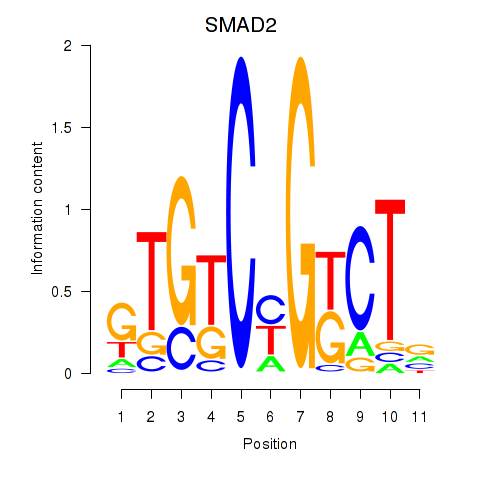
|
| TBX21_TBR1 | 0.664 |
|
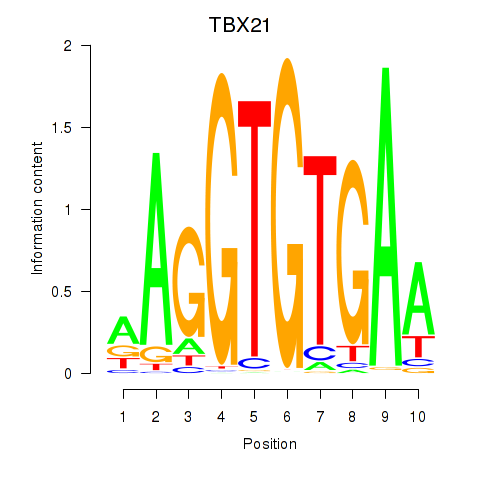
|
| ALX3 | 0.660 |
|
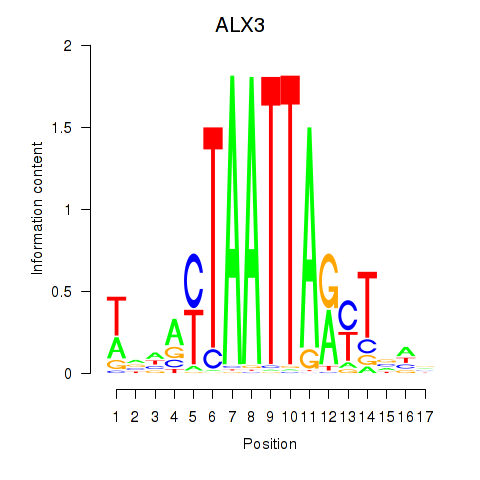
|
| GLIS3 | 0.658 |
|
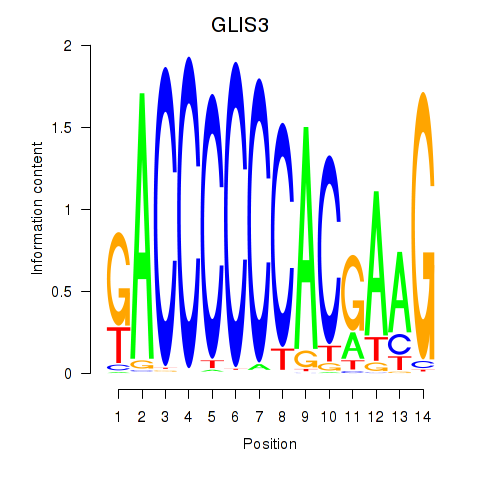
|
| PBX1 | 0.648 |
|
|
| TBP | 0.647 |
|
|
| ZNF263 | 0.629 |
|
|
| EVX2 | 0.603 |
|
|
| CPEB1 | 0.598 |
|
|
| NKX1-1 | 0.595 |
|
|
| ETV3 | 0.594 |
|
|
| ZFHX3 | 0.577 |
|
|
| E2F8 | 0.566 |
|
|
| EGR1_EGR4 | 0.556 |
|
|
| PRRX1_ALX4_PHOX2A | 0.554 |
|
|
| TBX2 | 0.546 |
|
|
| MAFA | 0.516 |
|
|
| ARNT | 0.516 |
|
|
| POU4F1_POU4F3 | 0.507 |
|
|
| MYOD1 | 0.506 |
|
|
| FOXD3_FOXI1_FOXF1 | 0.481 |
|
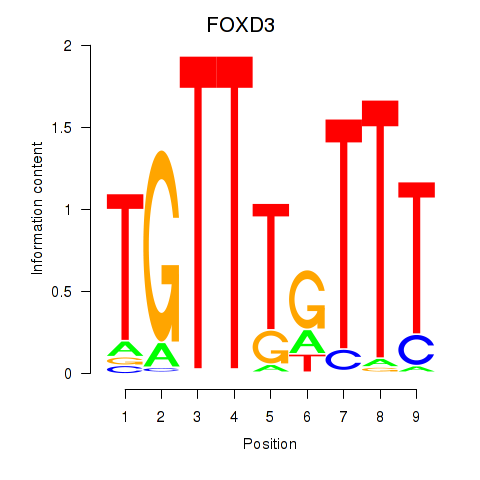
|
| ETS1 | 0.480 |
|
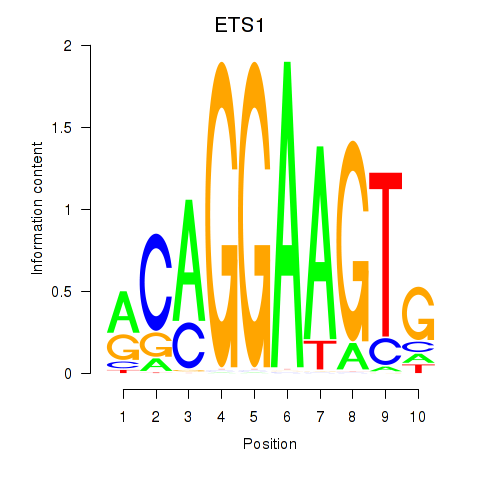
|
| POU3F2 | 0.477 |
|
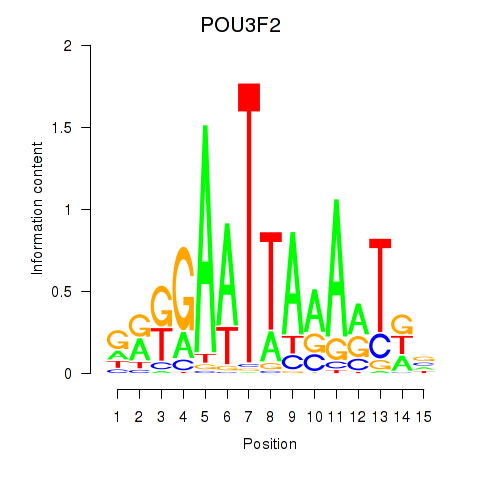
|
| FOXA1 | 0.460 |
|
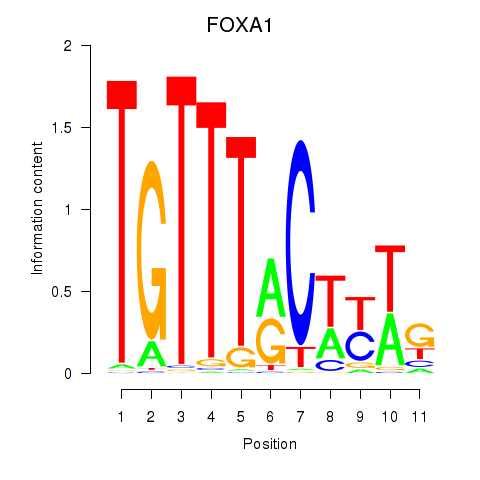
|
| BHLHE22_BHLHA15_BHLHE23 | 0.454 |
|
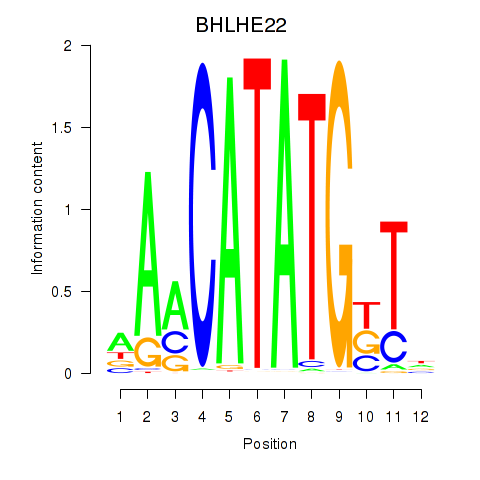
|
| NFATC1 | 0.446 |
|
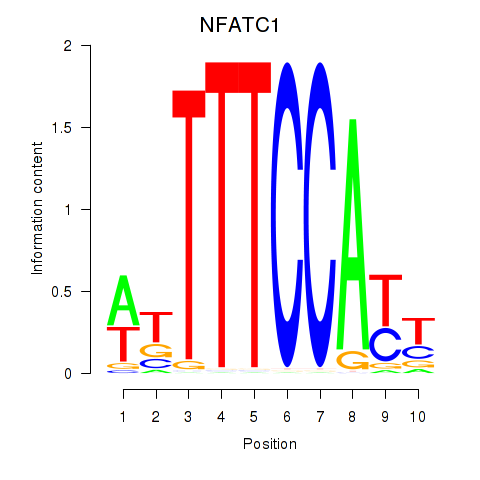
|
| BACH2 | 0.430 |
|
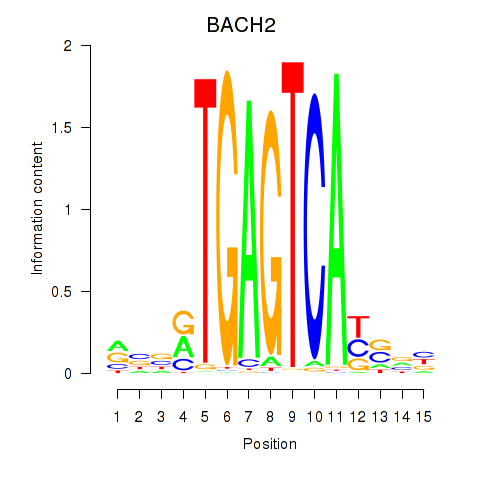
|
| LHX8 | 0.426 |
|
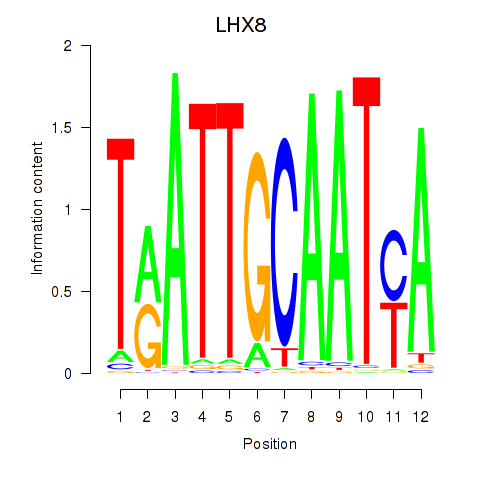
|
| PPARA | 0.426 |
|
|
| ZNF740_ZNF219 | 0.407 |
|
|
| MYF6 | 0.406 |
|
|
| LEF1 | 0.395 |
|
|
| ZBTB18 | 0.379 |
|
|
| GSC_GSC2 | 0.378 |
|
|
| FOSL1 | 0.373 |
|
|
| LMX1B_MNX1_RAX2 | 0.371 |
|
|
| HOXB2_UNCX_HOXD3 | 0.364 |
|
|
| RFX5 | 0.361 |
|
|
| HOXB13 | 0.352 |
|
|
| PLAG1 | 0.351 |
|
|
| MEF2B | 0.320 |
|
|
| HOXC11 | 0.315 |
|
|
| PBX2 | 0.313 |
|
|
| INSM1 | 0.295 |
|
|
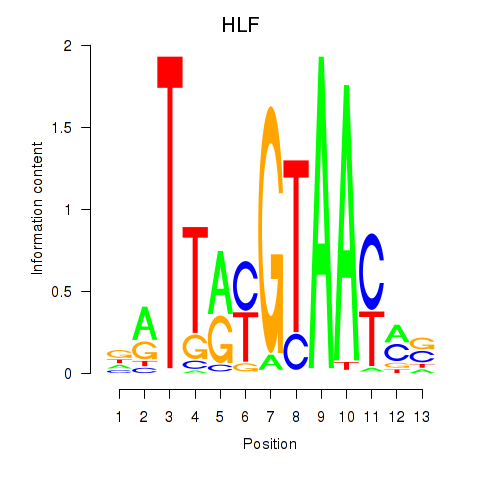
| HLF_TEF | 0.285 |
|
|
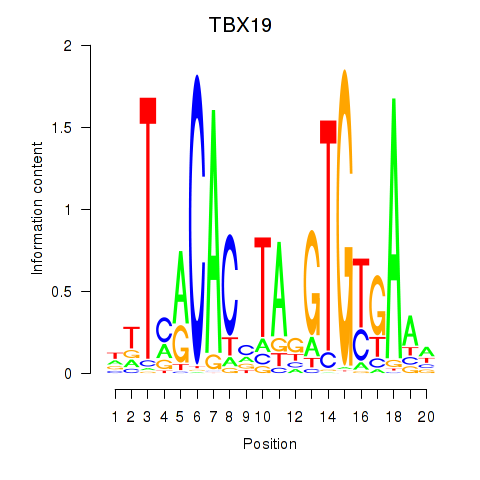
| TBX19 | 0.274 |
|
|
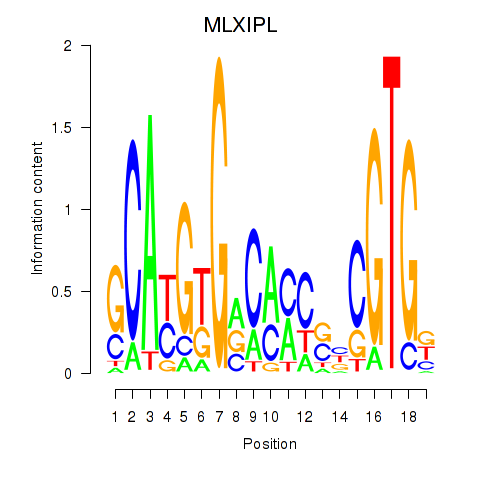
| MLXIPL | 0.262 |
|
|
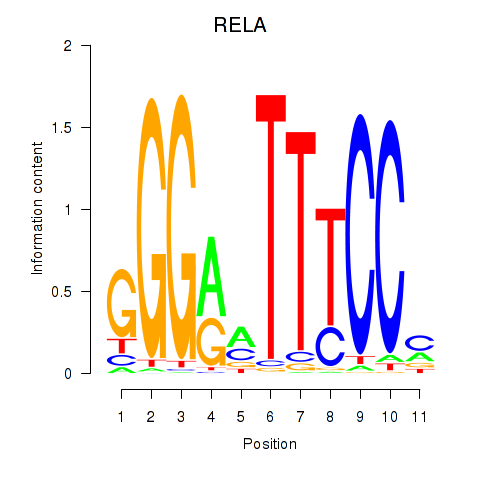
| RELA | 0.260 |
|
|
| AR_NR3C2 | 0.257 |
|
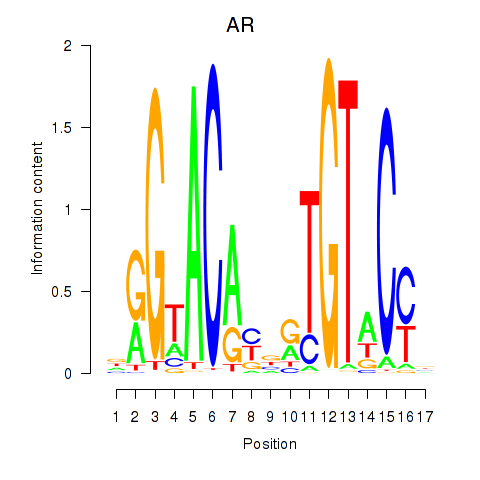
|
| HOXB4_LHX9 | 0.249 |
|
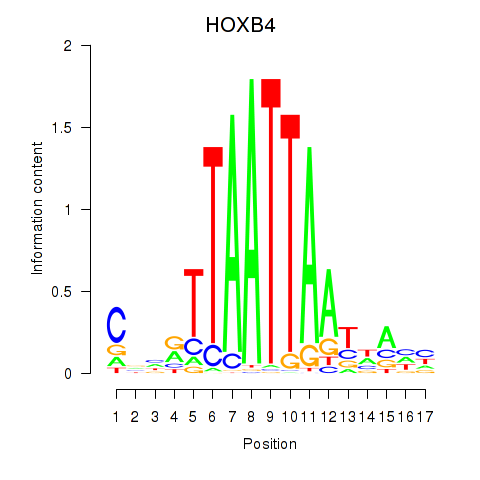
|
| GATA6 | 0.246 |
|
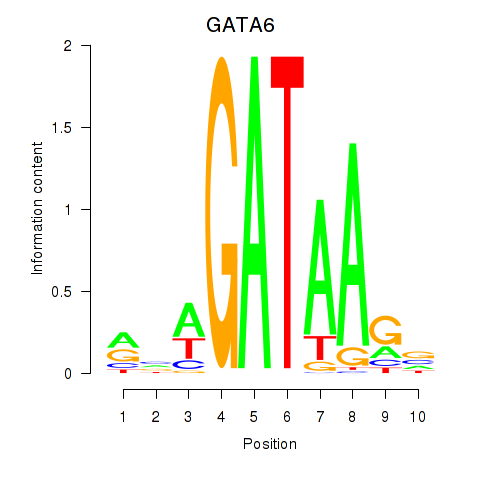
|
| HOXD1 | 0.237 |
|
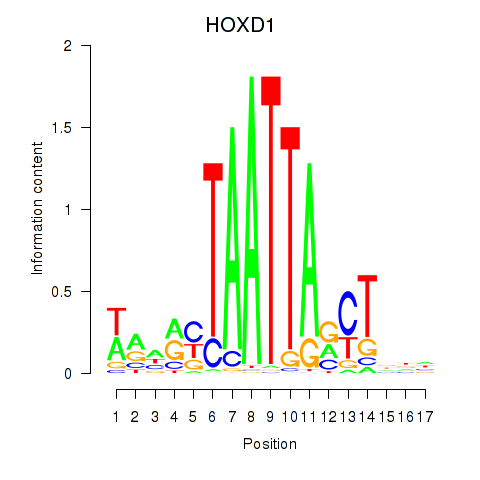
|
| ZBTB49 | 0.214 |
|
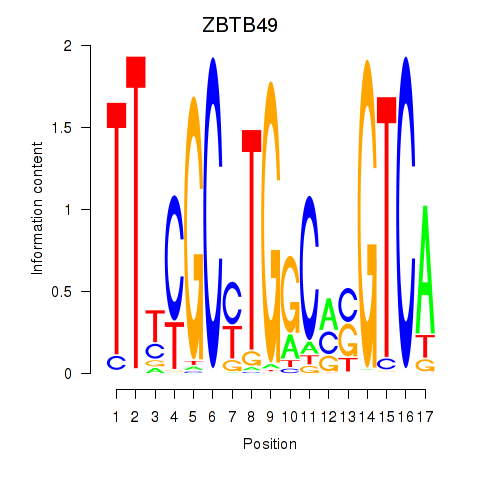
|
| NR4A1 | 0.206 |
|
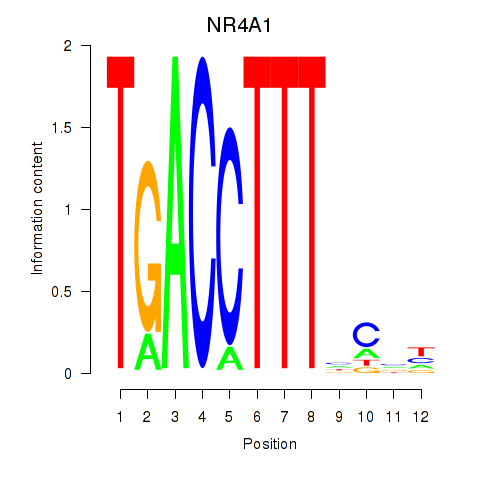
|
| SREBF2 | 0.203 |
|
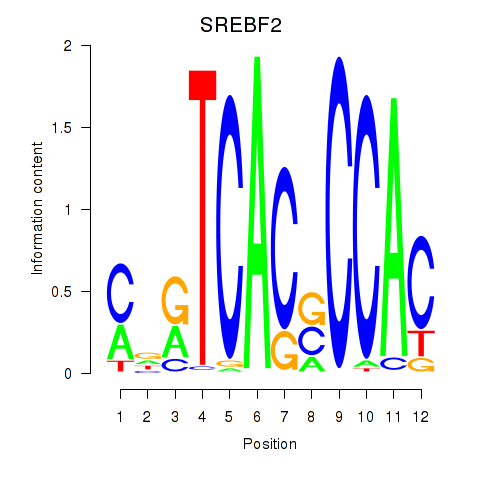
|
| T | 0.202 |
|
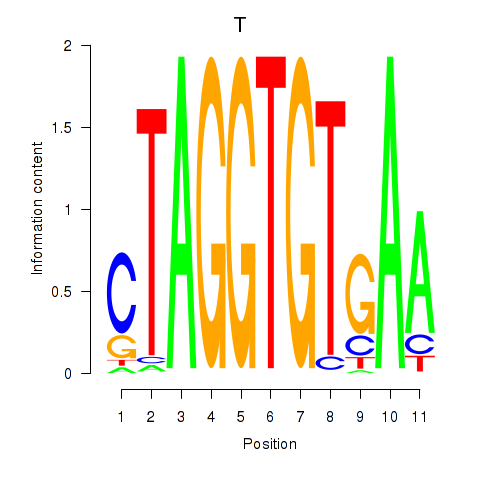
|
| NFATC4 | 0.194 |
|
|
| OLIG1 | 0.190 |
|
|
| NKX2-4 | 0.148 |
|
|
| HOXA10_HOXB9 | 0.129 |
|
|
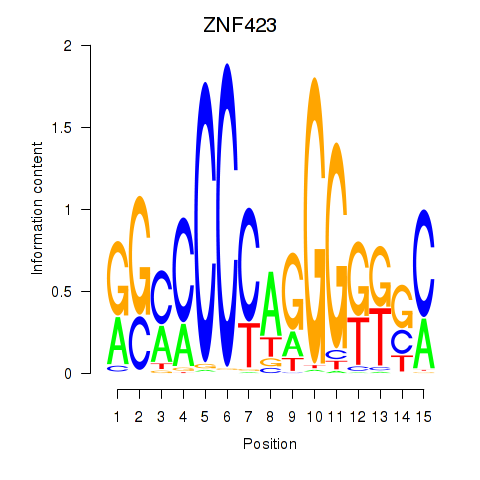
| ZNF423 | 0.122 |
|
|
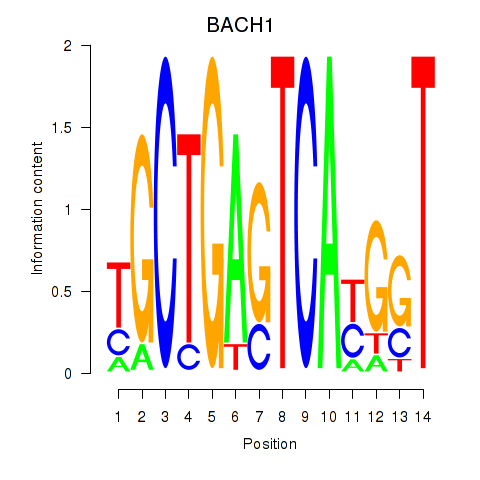
| BACH1_NFE2_NFE2L2 | 0.110 |
|
|
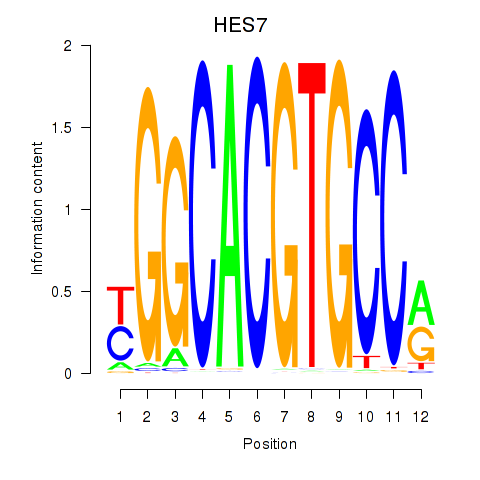
| HES7_HES5 | 0.109 |
|
|
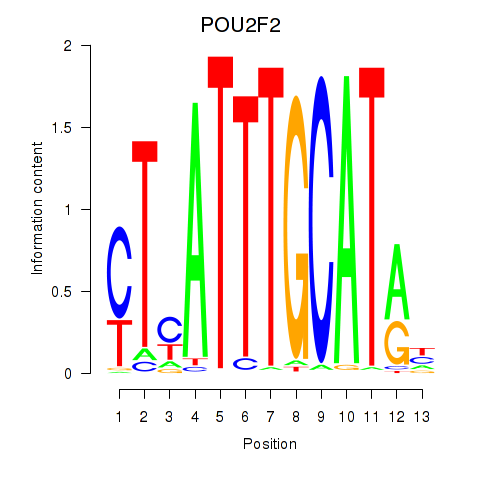
| POU2F2_POU3F1 | 0.107 |
|
|
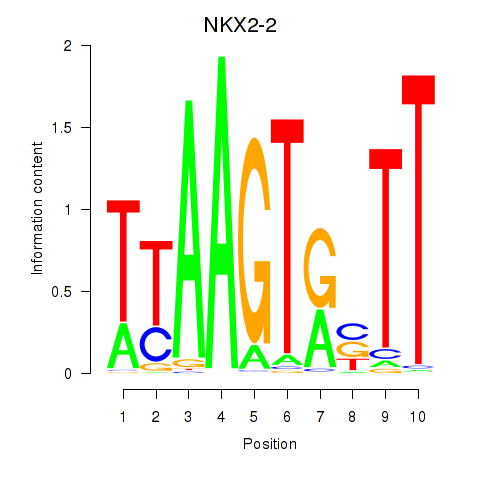
| NKX2-2 | 0.103 |
|
|
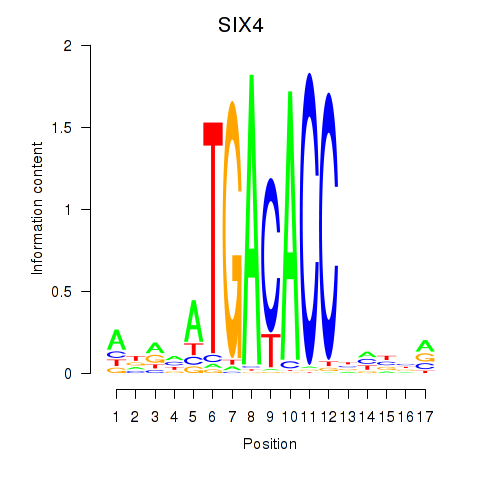
| SIX4 | 0.097 |
|
|
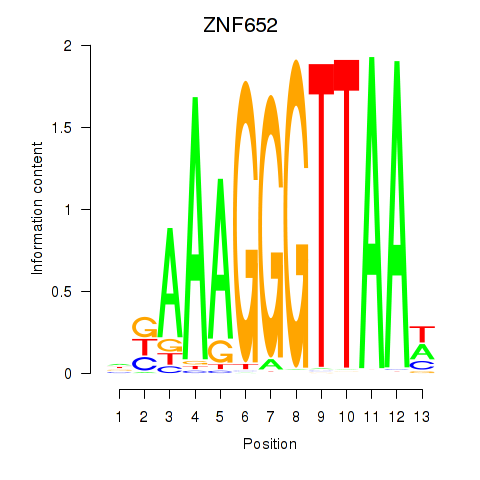
| ZNF652 | 0.095 |
|
|
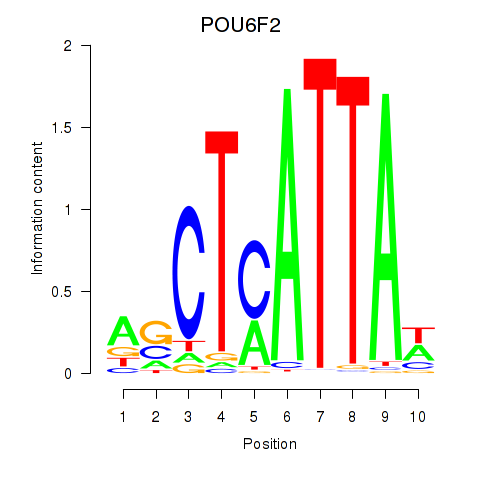
| POU6F2 | 0.092 |
|
|
| ZNF333 | 0.092 |
|
|
| EMX1 | 0.091 |
|
|
| NKX1-2_RAX | 0.090 |
|
|
| VDR | 0.086 |
|
|
| SIX6 | 0.083 |
|
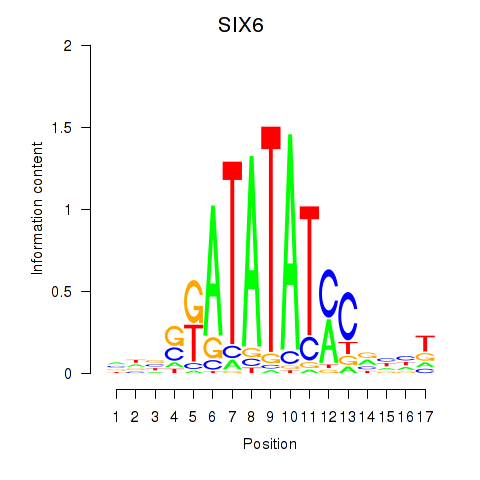
|
| NFAT5 | 0.070 |
|
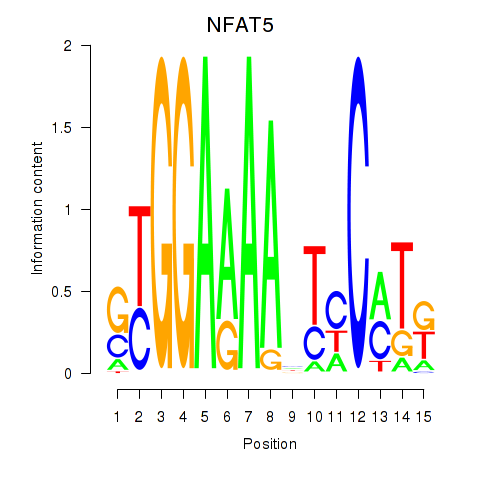
|
| BARHL2 | 0.070 |
|
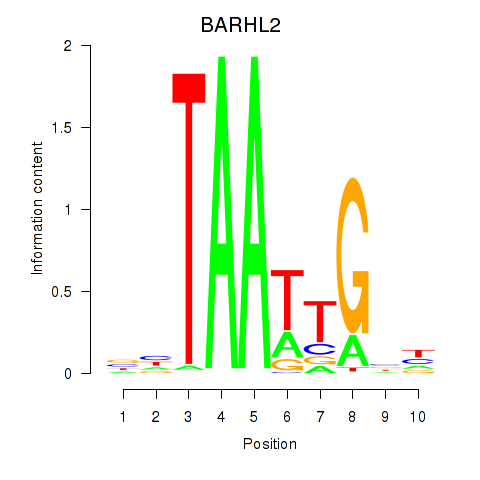
|
| DPRX | 0.068 |
|
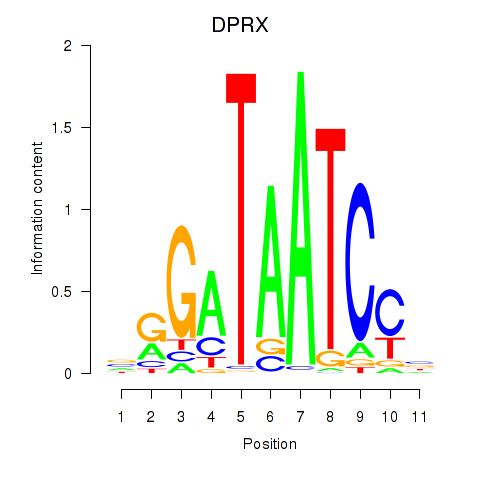
|
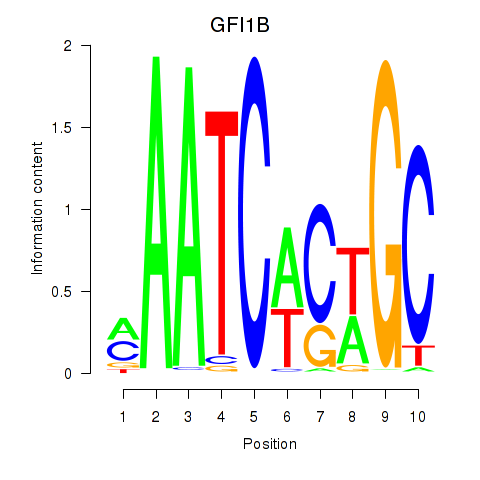
| GFI1B | 0.065 |
|
|
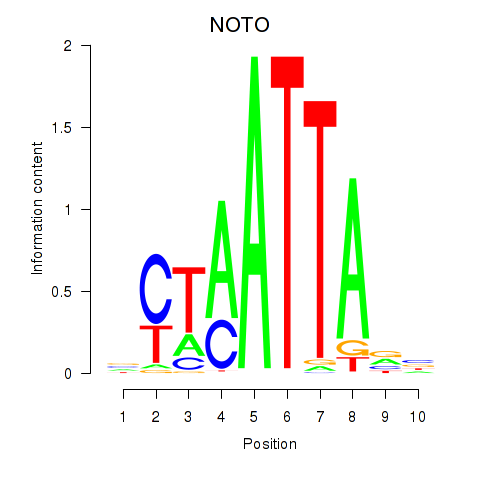
| NOTO_VSX2_DLX2_DLX6_NKX6-1 | 0.056 |
|
|
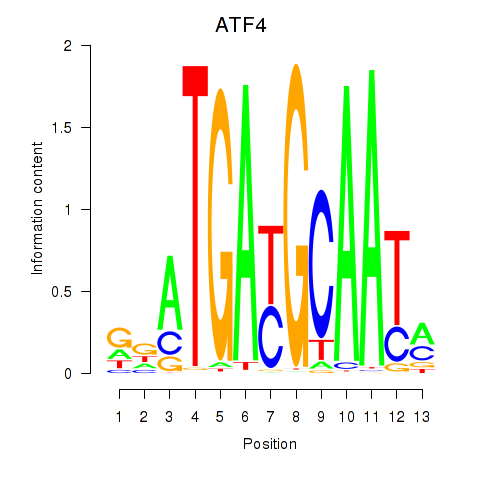
| ATF4 | 0.054 |
|
|
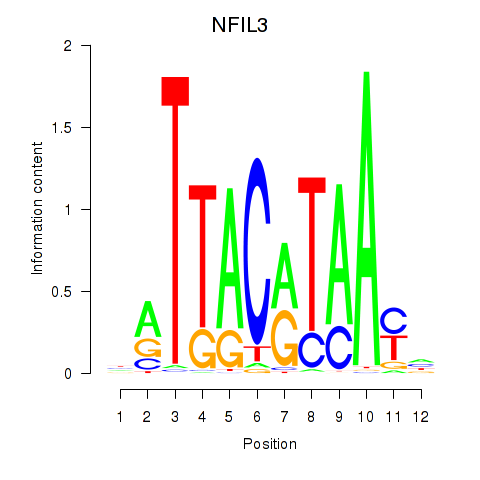
| NFIL3 | 0.047 |
|
|
| TFEC_MITF_ARNTL_BHLHE41 | 0.045 |
|
|
| PITX2 | 0.044 |
|
|
| NR2C1 | 0.042 |
|
|
| GLIS1 | 0.018 |
|
|
| BATF3 | 0.013 |
|
|
| HOXB8 | 0.010 |
|
|
| VAX1_GSX2 | 0.005 |
|
|
| E4F1 | 0.000 |
|
|
| EZH2 | -0.000 |
|
|
| HOXA4 | 0.000 |
|
|
| HOXA6 | -0.000 |
|
|
| HOXB6_PRRX2 | 0.000 |
|
|
| KLF6 | -0.000 |
|
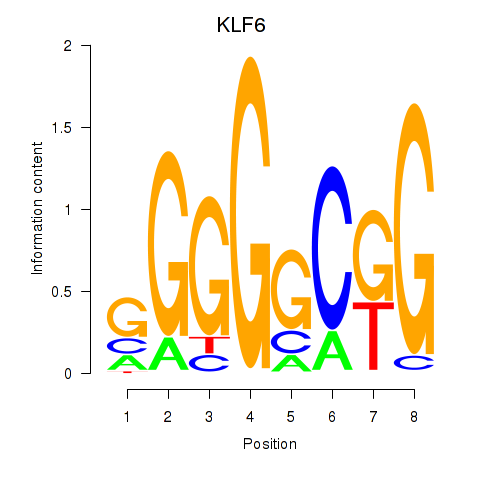
|
| SPI1 | -0.000 |
|
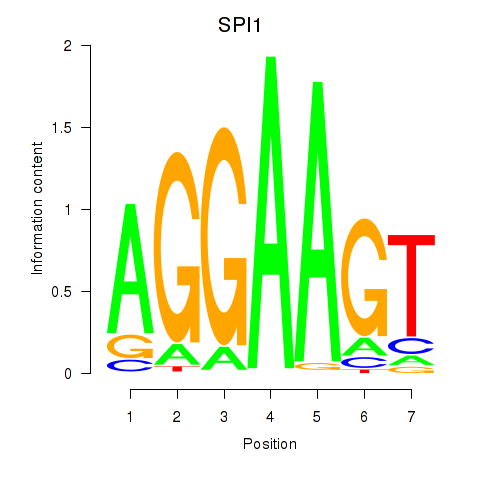
|
| TFDP1 | -0.000 |
|
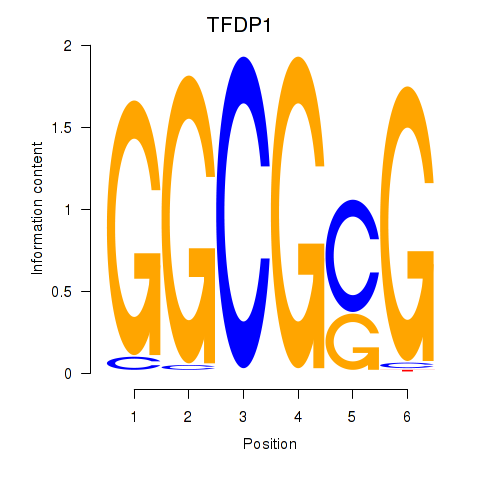
|
| CDX2 | -0.001 |
|
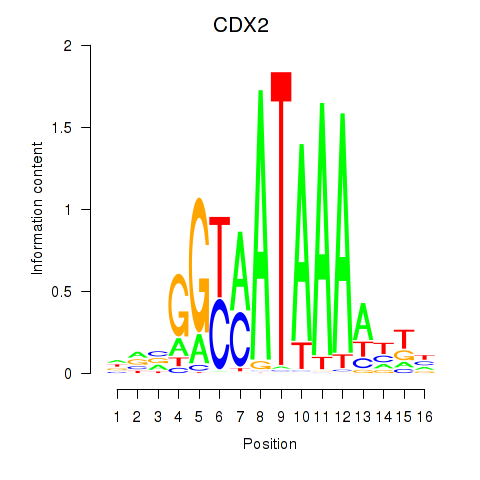
|
| EN2_GBX2_LBX2 | -0.002 |
|
|
| CBFB | -0.005 |
|
|
| ZNF350 | -0.012 |
|
|
| STAT4 | -0.013 |
|
|
| BHLHE40 | -0.014 |
|
|
| EBF1 | -0.014 |
|
|
| ZBTB6 | -0.024 |
|
|
| ETV2 | -0.027 |
|
|
| FOXO4 | -0.028 |
|
|
| NKX6-3 | -0.031 |
|
|
| BCL6B | -0.037 |
|
|
| HOXD4 | -0.043 |
|
|
| DBP | -0.049 |
|
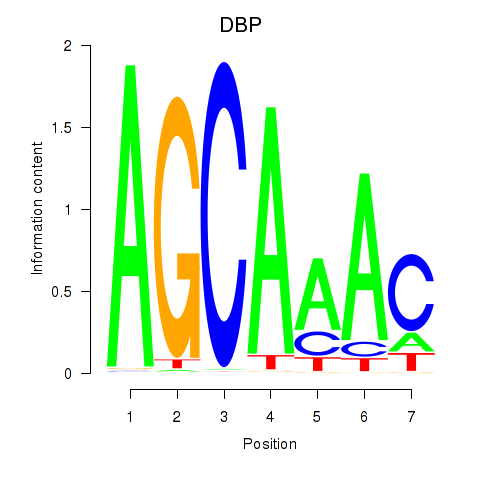
|
| ZBED1 | -0.055 |
|
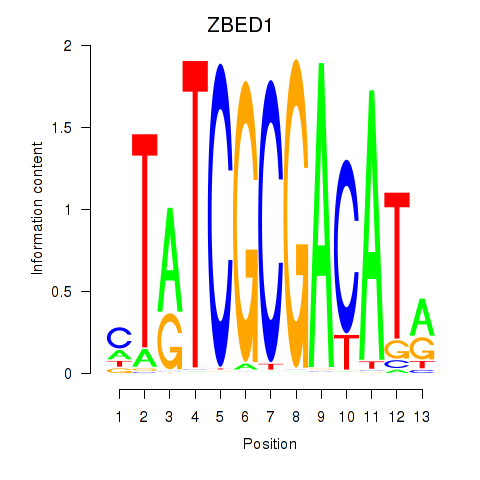
|
| E2F7_E2F1 | -0.057 |
|
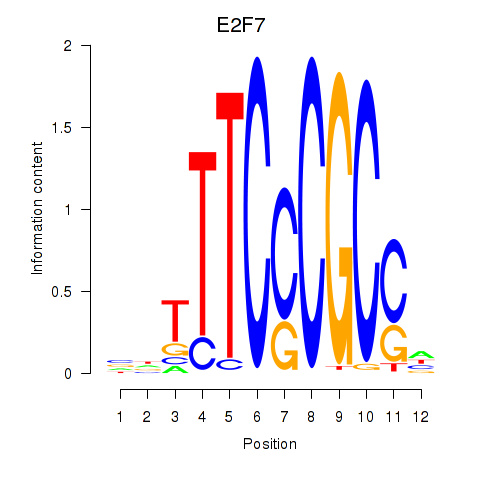
|
| KLF15 | -0.061 |
|
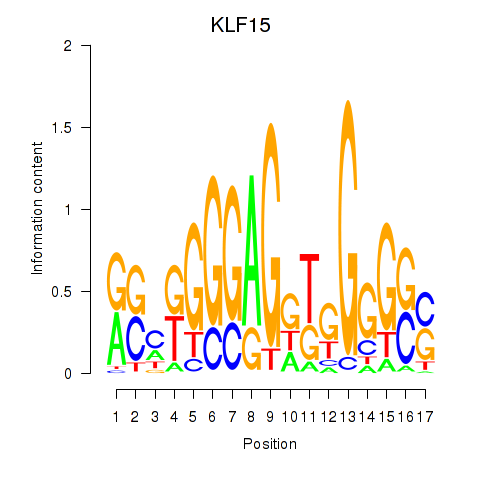
|
| TBX4 | -0.064 |
|
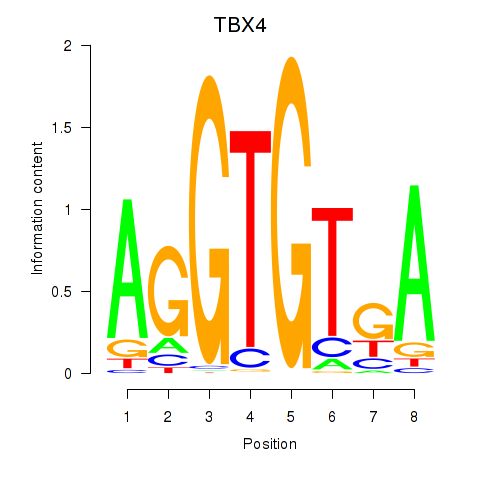
|
| PPARG | -0.064 |
|
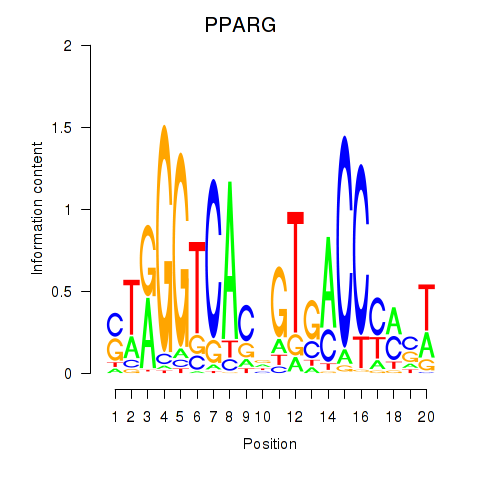
|
| NR1I3 | -0.072 |
|
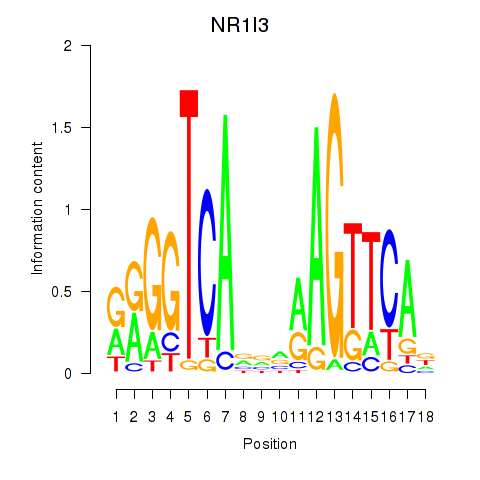
|
| PDX1 | -0.083 |
|
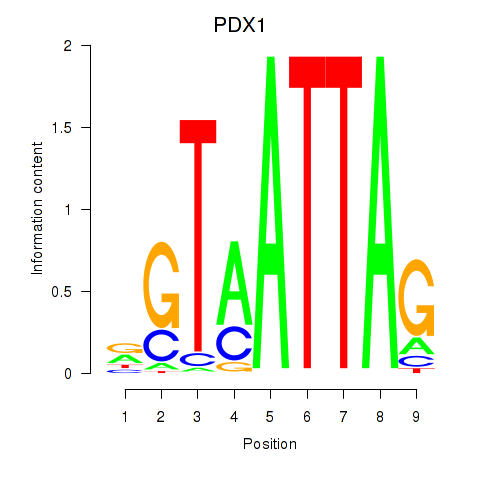
|
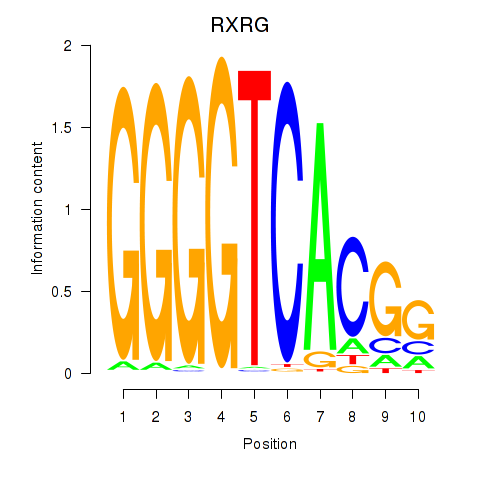
| RXRG | -0.092 |
|
|
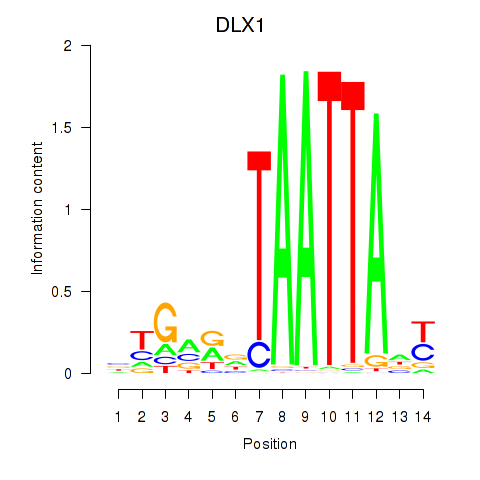
| DLX1_HOXA3_BARX2 | -0.096 |
|
|
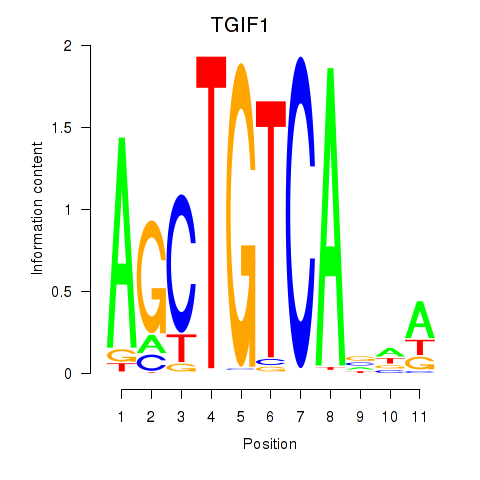
| TGIF1 | -0.098 |
|
|
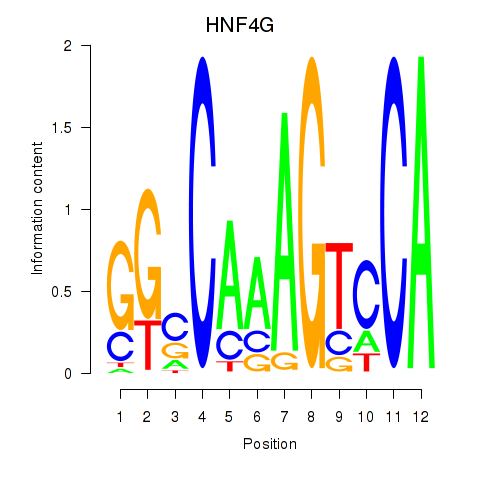
| HNF4G | -0.106 |
|
|
| RBPJ | -0.123 |
|
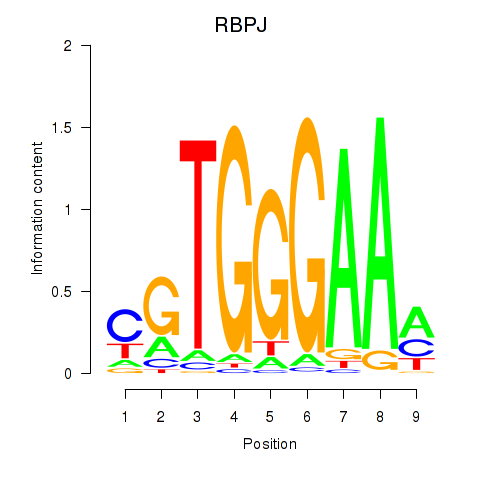
|
| HMX3 | -0.124 |
|
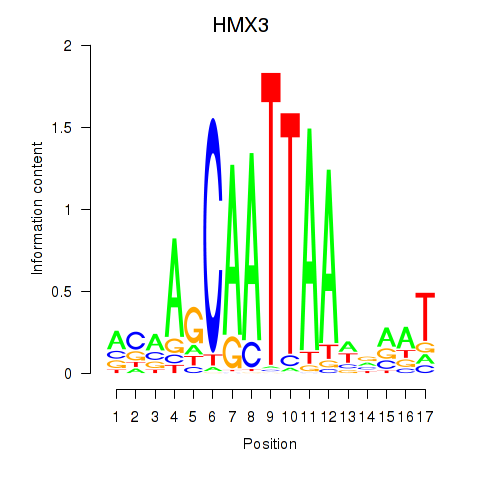
|
| GCM1 | -0.127 |
|
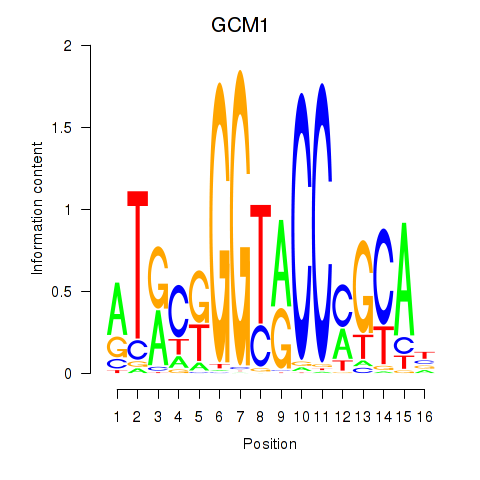
|
| FUBP1 | -0.130 |
|
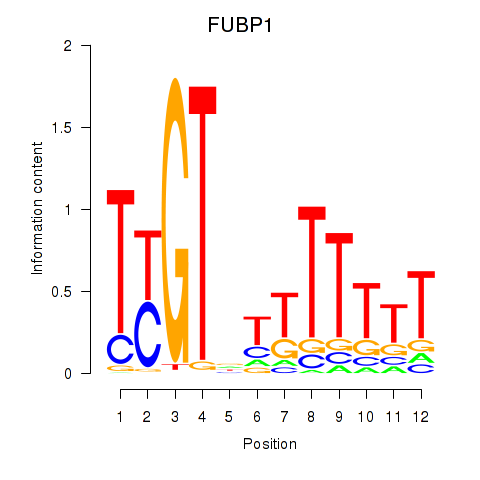
|
| DMC1 | -0.134 |
|
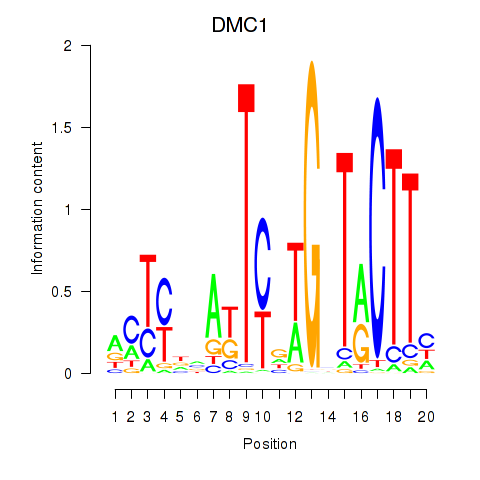
|
| HINFP1 | -0.136 |
|
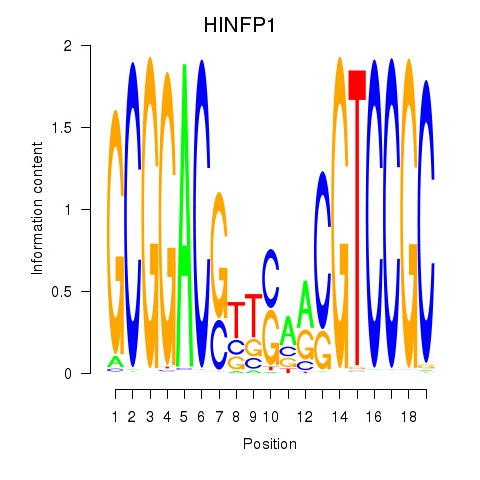
|
| TCF7L2 | -0.140 |
|
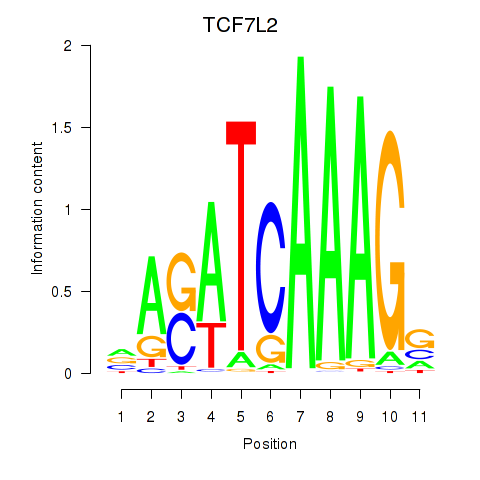
|
| ZKSCAN1 | -0.142 |
|
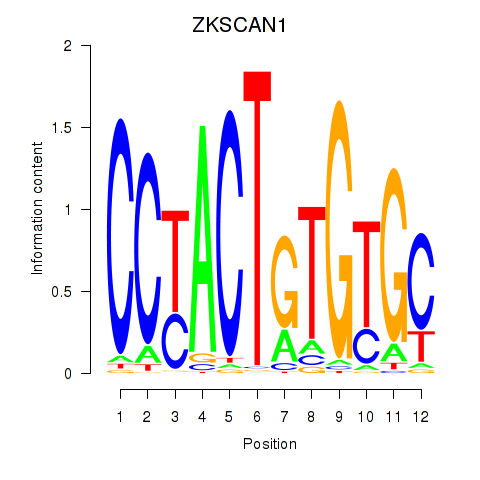
|
| HOXD11_HOXA11 | -0.159 |
|
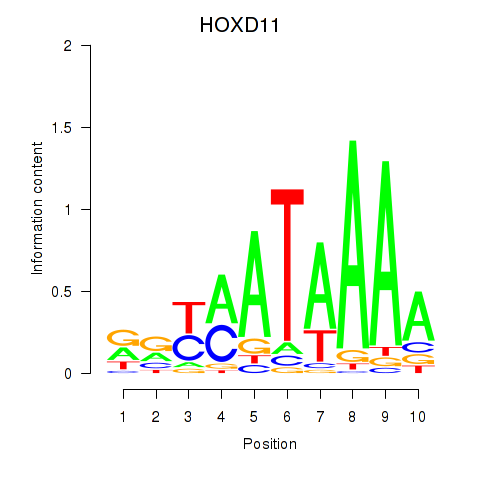
|
| GLIS2 | -0.164 |
|
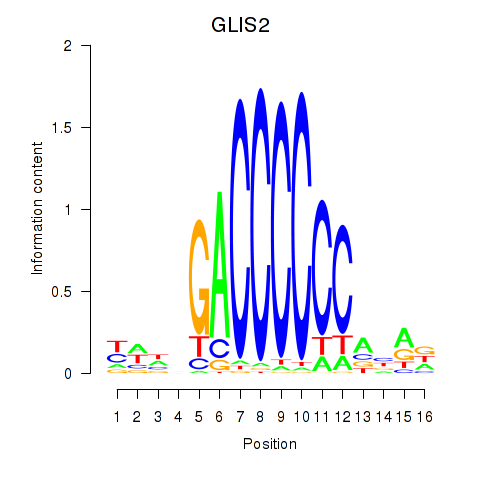
|
| PTF1A | -0.168 |
|
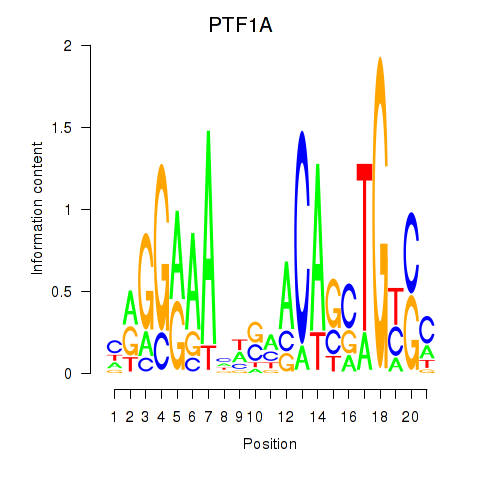
|
| ZNF35 | -0.169 |
|
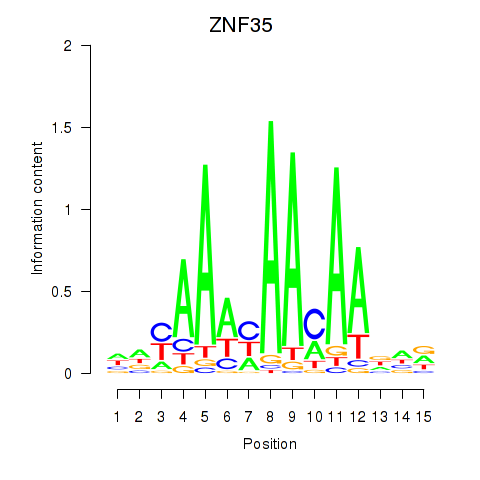
|
| FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 | -0.178 |
|
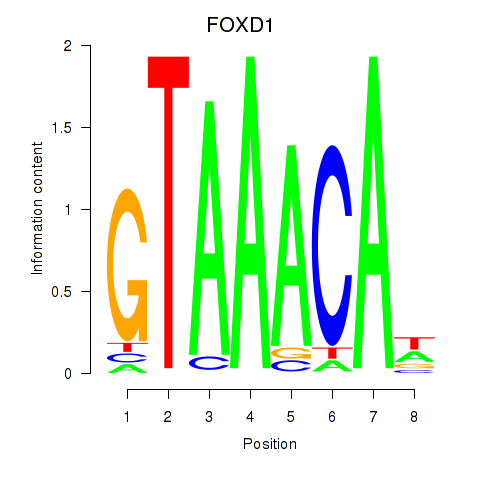
|
| BBX | -0.178 |
|
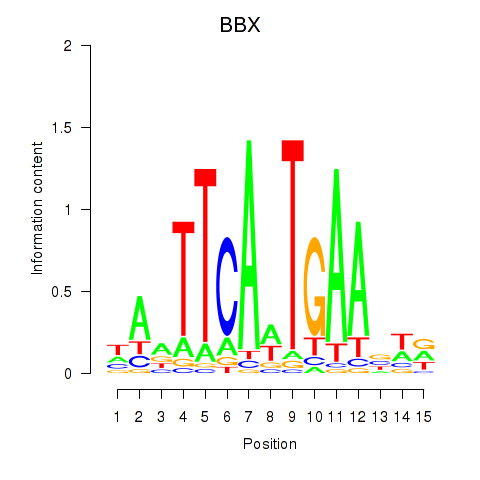
|
| SOX5 | -0.189 |
|
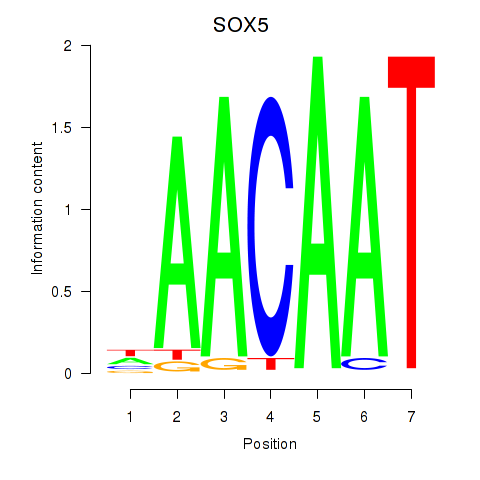
|
| TFAP2E | -0.192 |
|
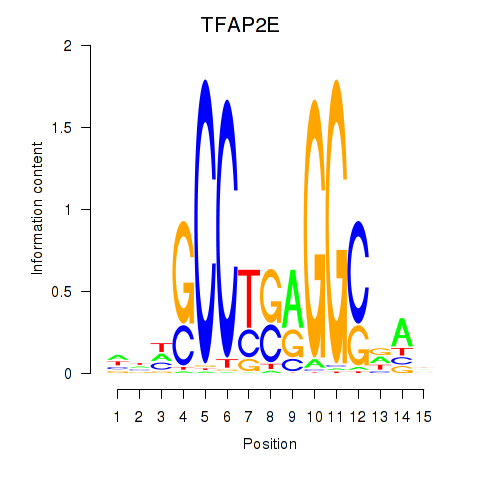
|
| PKNOX2 | -0.194 |
|
|
| HOXB7 | -0.198 |
|
|
| OTX2_CRX | -0.207 |
|
|
| KLF7 | -0.213 |
|
|
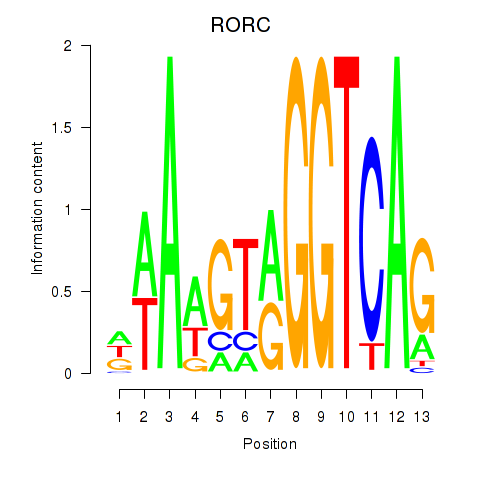
| RORC | -0.218 |
|
|
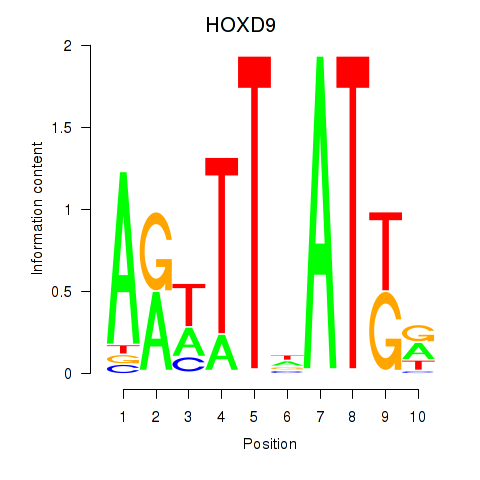
| HOXD9 | -0.220 |
|
|
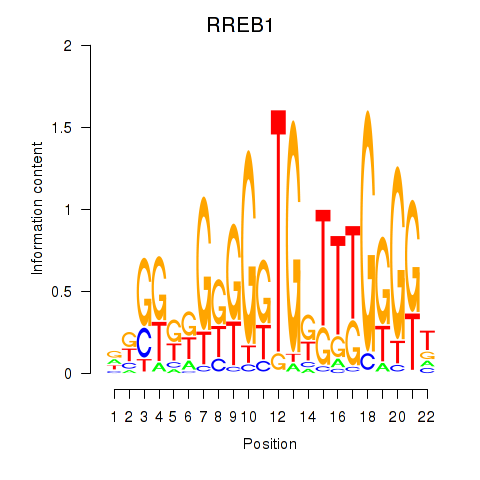
| RREB1 | -0.221 |
|
|
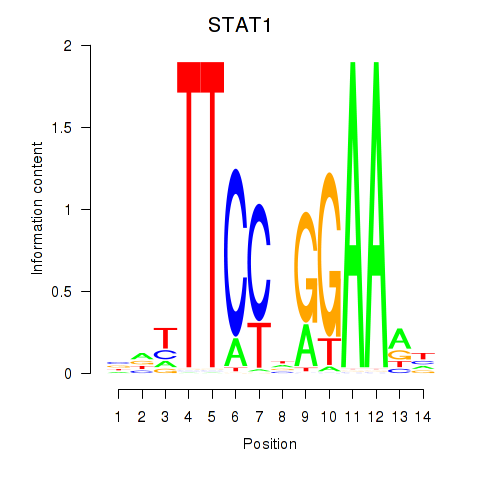
| STAT1_STAT3_BCL6 | -0.242 |
|
|
| SPDEF | -0.254 |
|
|
| GZF1 | -0.261 |
|
|
| STAT5B | -0.267 |
|
|
| STAT6 | -0.269 |
|
|
| EBF3 | -0.277 |
|
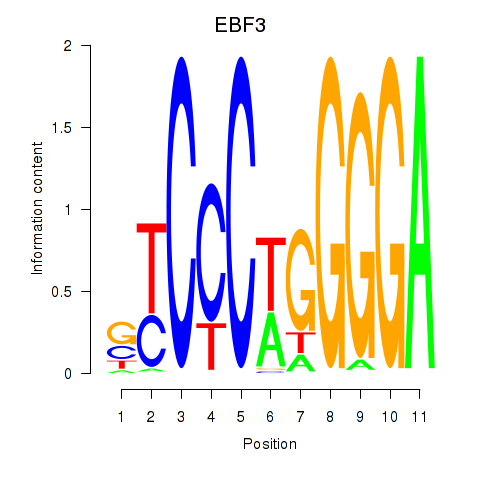
|
| E2F2_E2F5 | -0.277 |
|
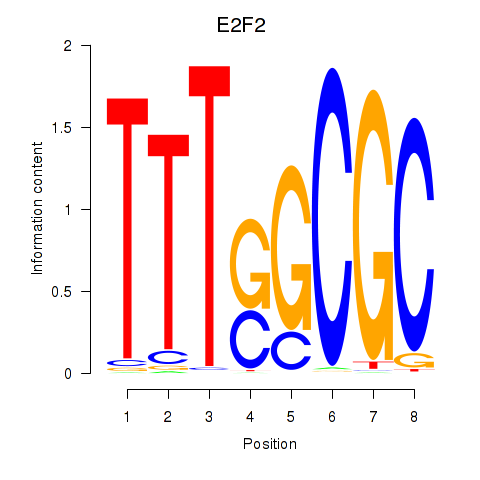
|
| HINFP | -0.285 |
|
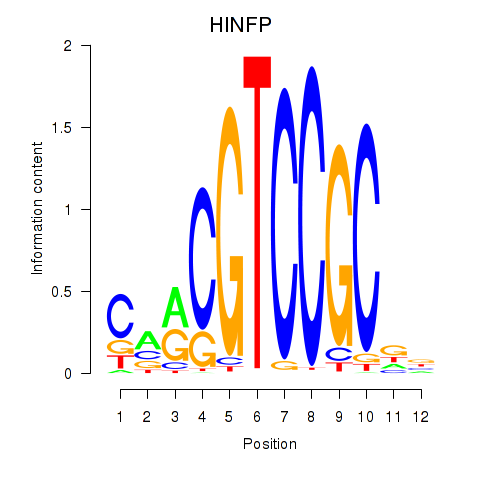
|
| CLOCK | -0.294 |
|
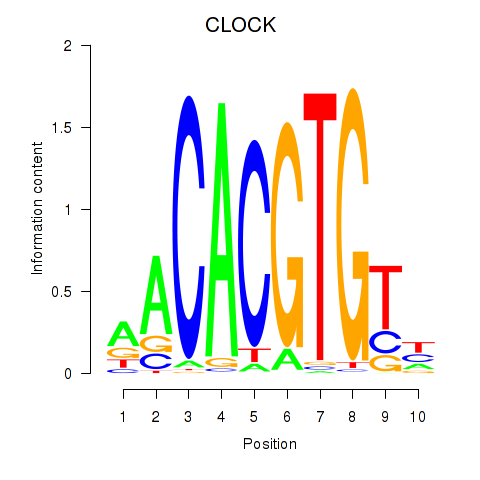
|
| SOX14 | -0.304 |
|
|
| DMBX1 | -0.304 |
|
|
| CREB1 | -0.305 |
|
|
| FLI1 | -0.314 |
|
|
| MLX_USF2_USF1_PAX2 | -0.316 |
|
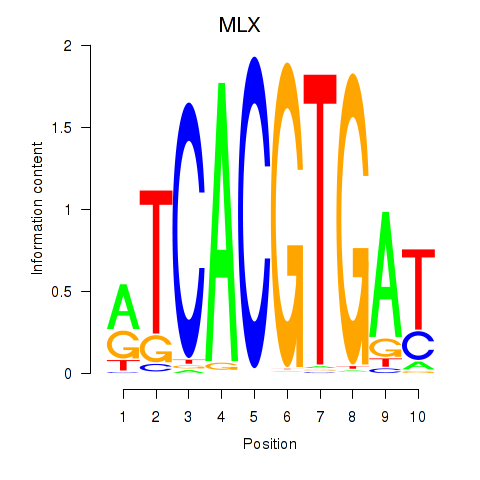
|
| SOX30 | -0.328 |
|
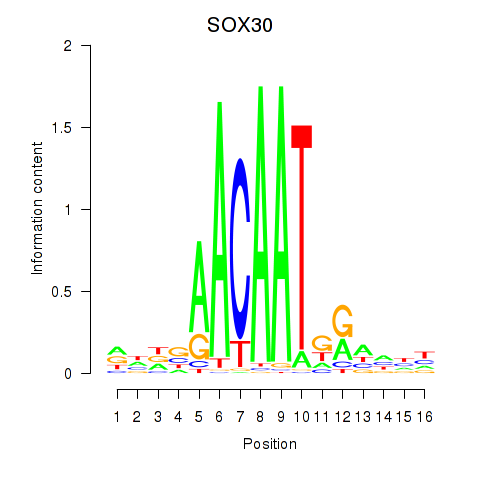
|
| ZNF410 | -0.348 |
|
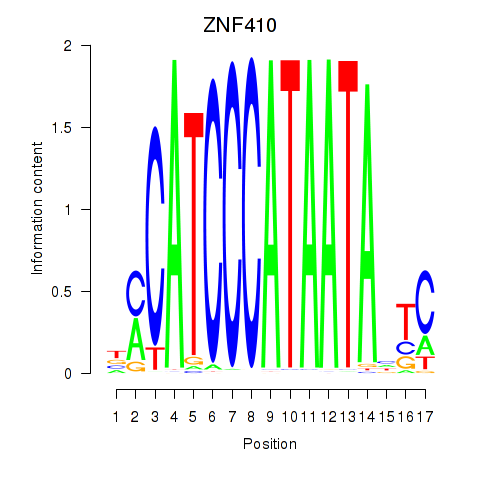
|
| FOSB | -0.353 |
|
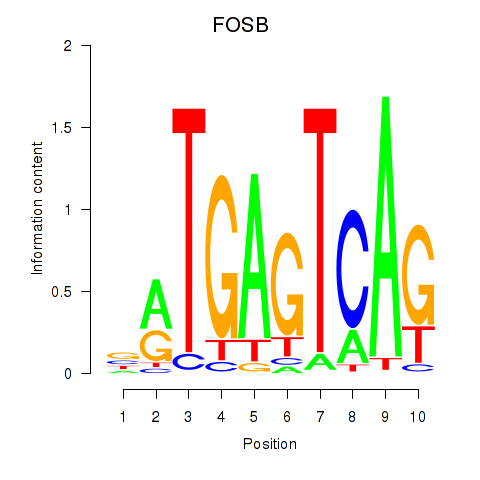
|
| STAT5A | -0.354 |
|
|
| NKX2-3 | -0.354 |
|
|
| FIGLA | -0.354 |
|
|
| TCF3_MYOG | -0.357 |
|
|
| NKX3-1 | -0.391 |
|
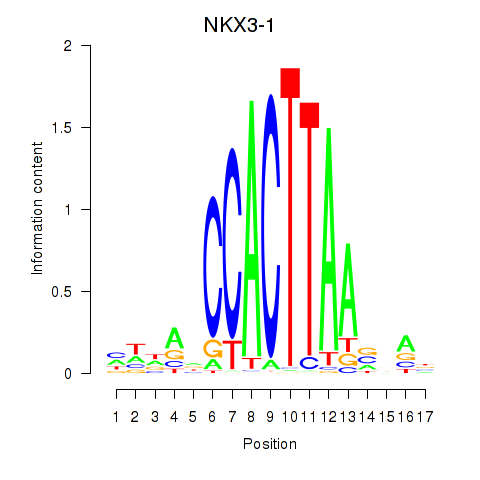
|
| REL | -0.415 |
|
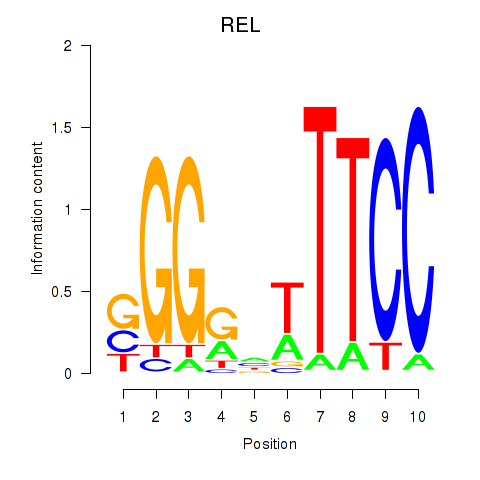
|
| SRY | -0.417 |
|
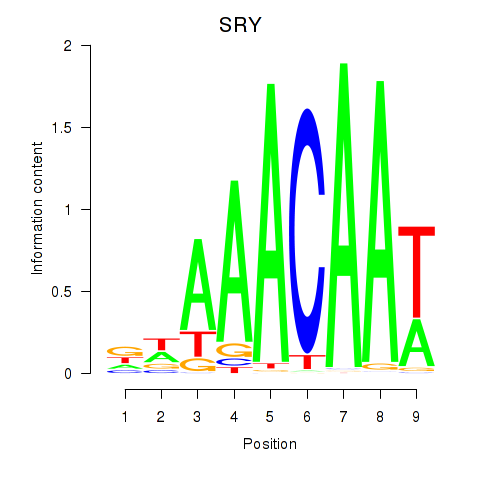
|
| PPARD | -0.426 |
|
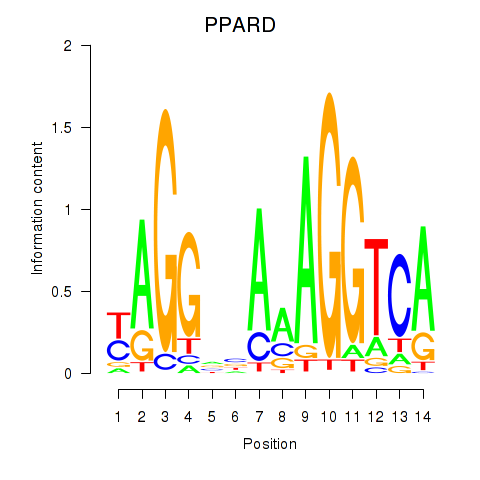
|
| SRF | -0.427 |
|
|
| TBX20 | -0.430 |
|
|
| SOX10_SOX15 | -0.432 |
|
|
| IRF3 | -0.446 |
|
|
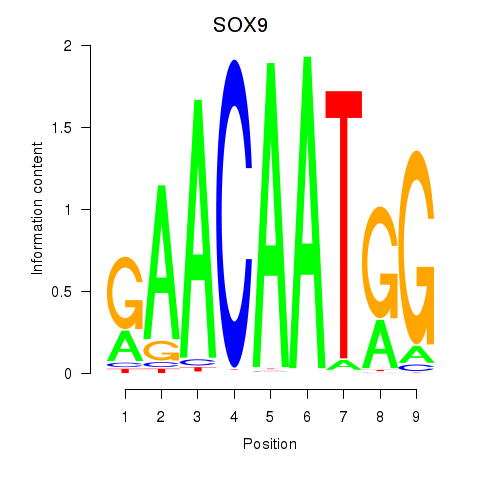
| SOX9 | -0.446 |
|
|
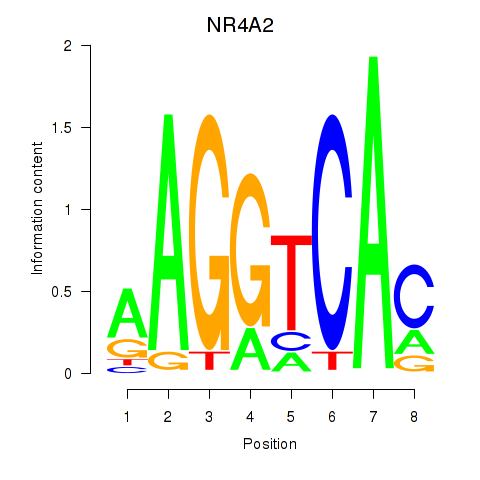
| NR4A2 | -0.447 |
|
|
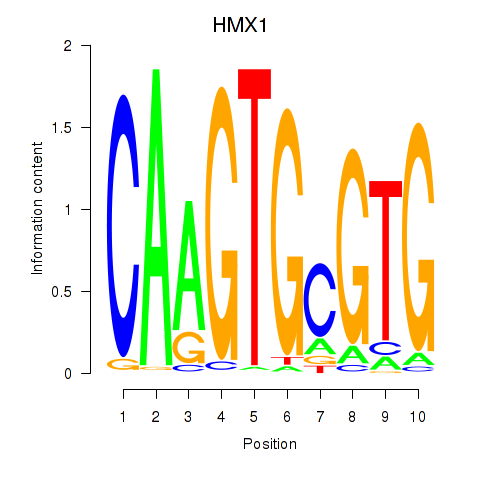
| HMX1 | -0.475 |
|
|
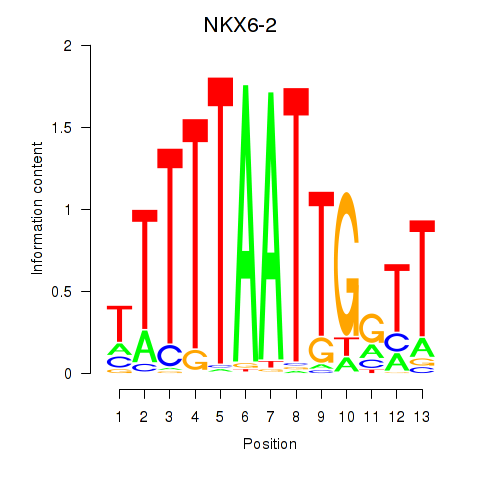
| NKX6-2 | -0.479 |
|
|
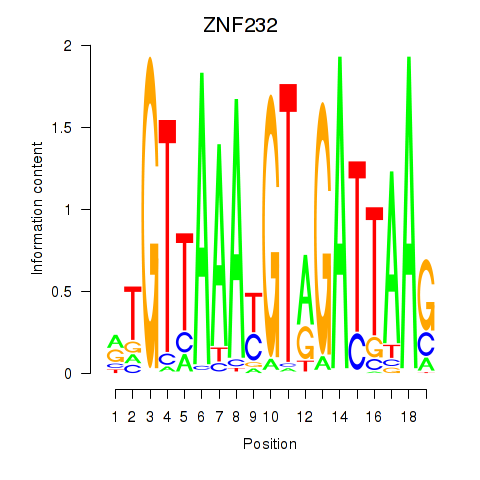
| ZNF232 | -0.481 |
|
|
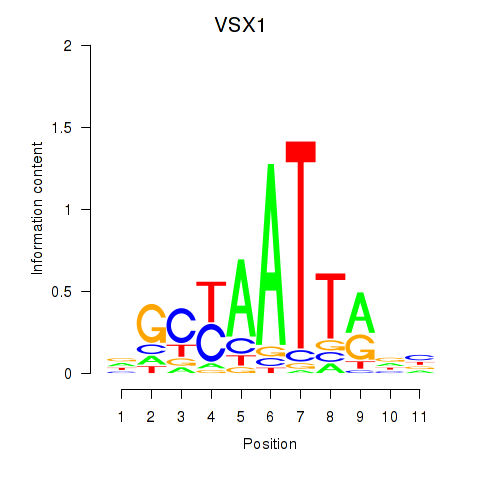
| VSX1 | -0.481 |
|
|
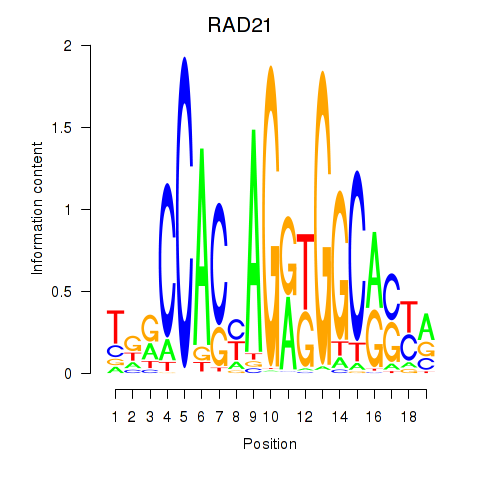
| RAD21_SMC3 | -0.482 |
|
|
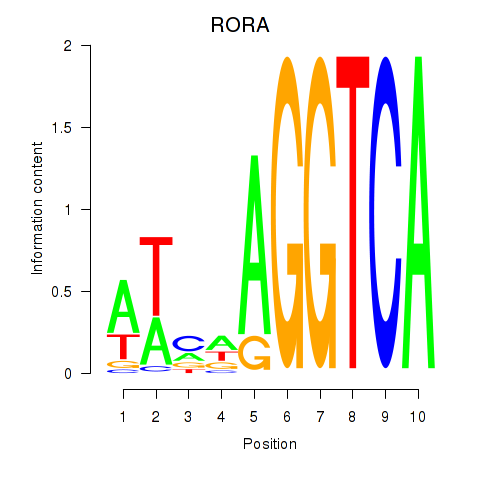
| RORA | -0.486 |
|
|
| CREB3L2 | -0.492 |
|
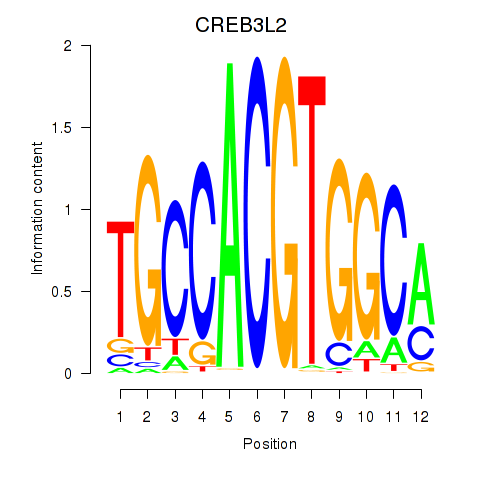
|
| OVOL1 | -0.502 |
|
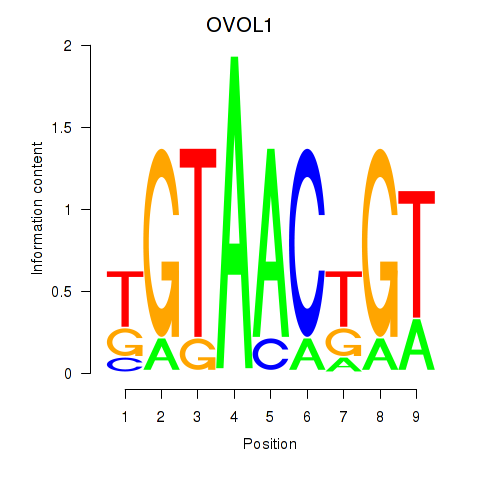
|
| SCRT1_SCRT2 | -0.514 |
|
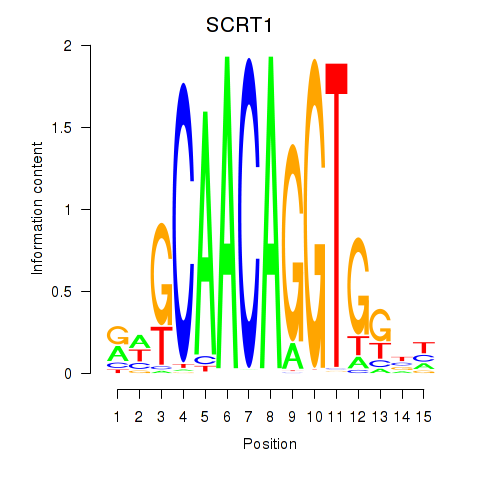
|
| FOXA3_FOXC2 | -0.515 |
|
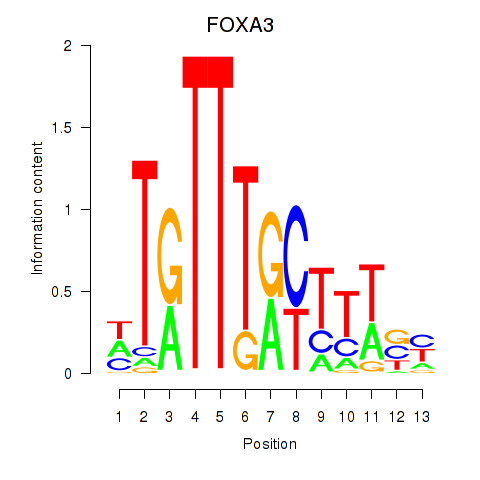
|
| HSF2 | -0.519 |
|
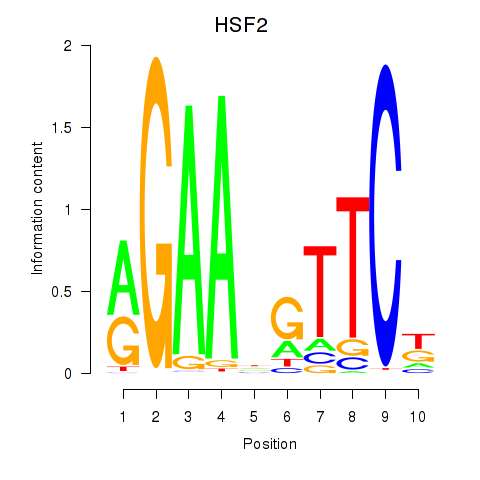
|
| SOX1 | -0.534 |
|
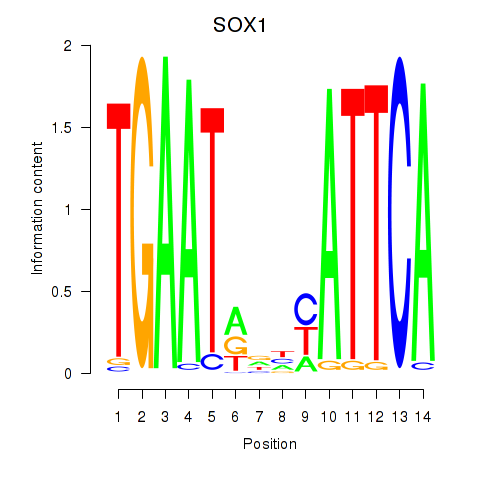
|
| CEBPE_CEBPD | -0.535 |
|
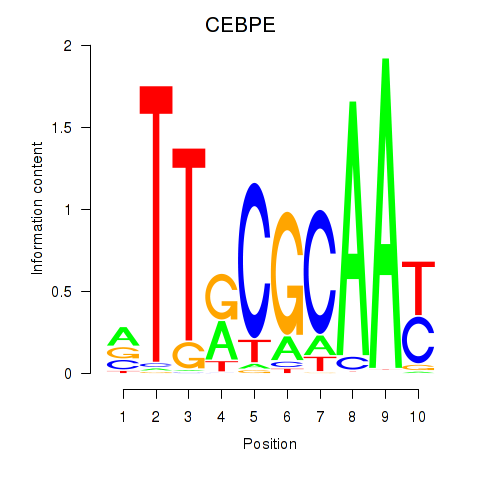
|
| CDX1 | -0.539 |
|
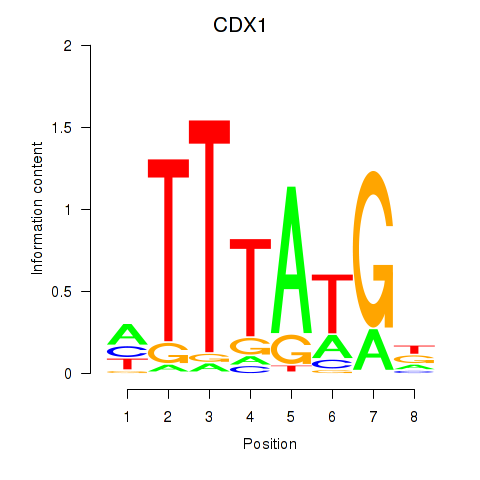
|
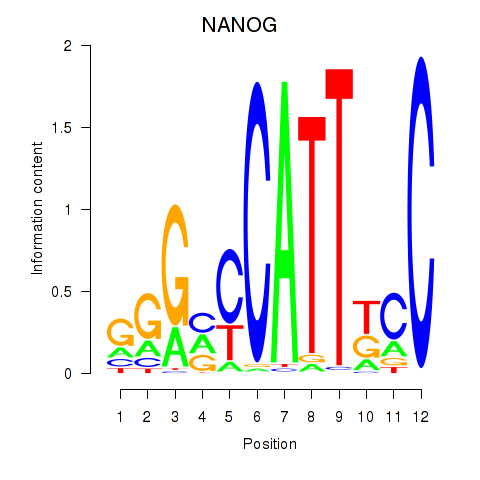
| NANOG | -0.540 |
|
|
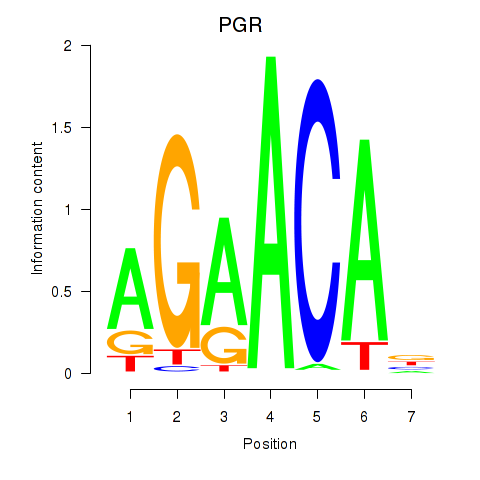
| PGR | -0.550 |
|
|
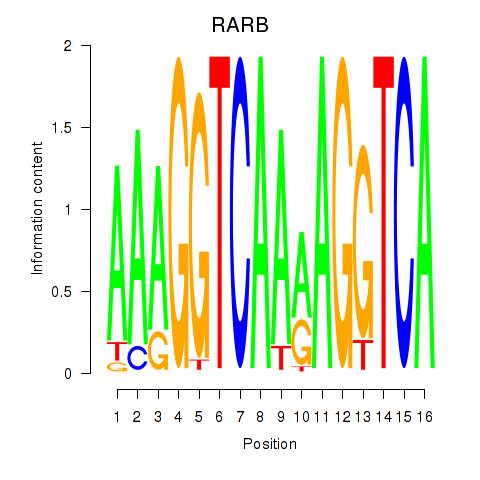
| RARB | -0.572 |
|
|
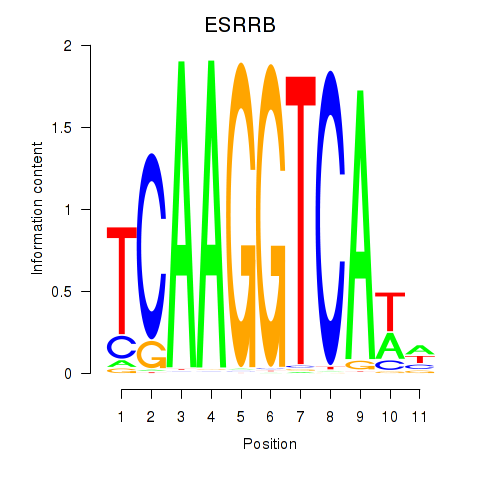
| ESRRB_ESRRG | -0.572 |
|
|
| MEF2C | -0.578 |
|
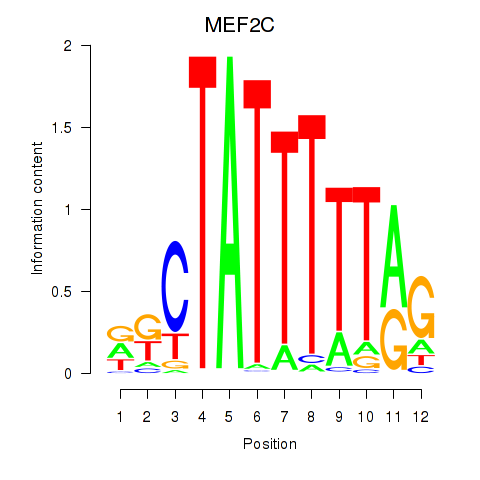
|
| HOXC9 | -0.584 |
|
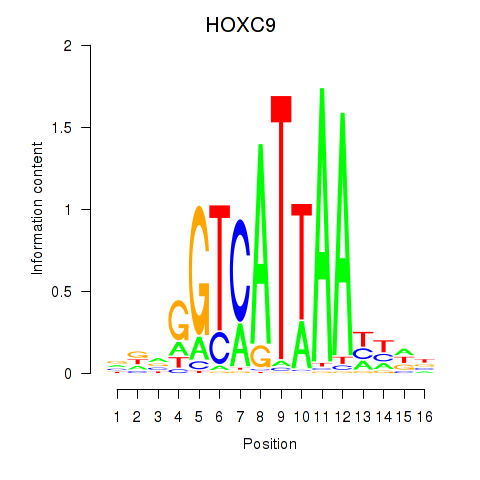
|
| NFIA | -0.590 |
|
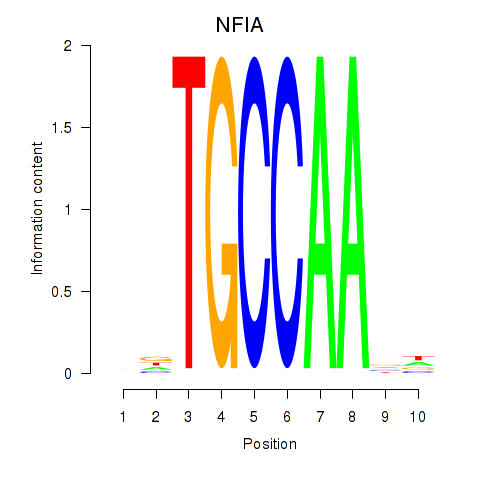
|
| ETV4_ETS2 | -0.602 |
|
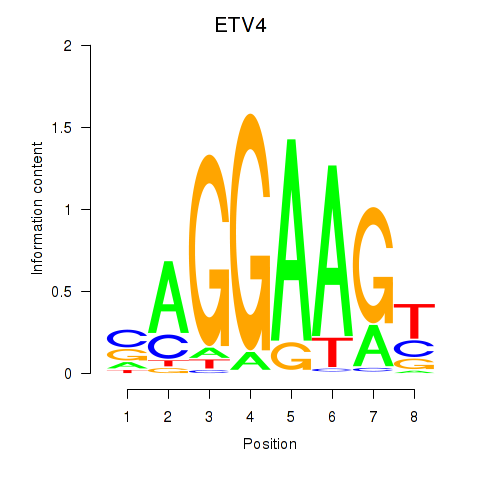
|
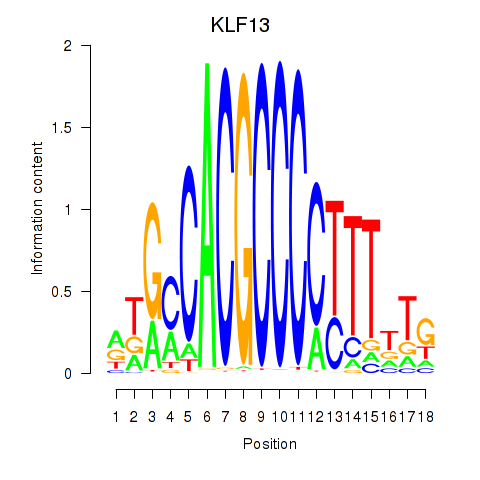
| KLF13 | -0.630 |
|
|
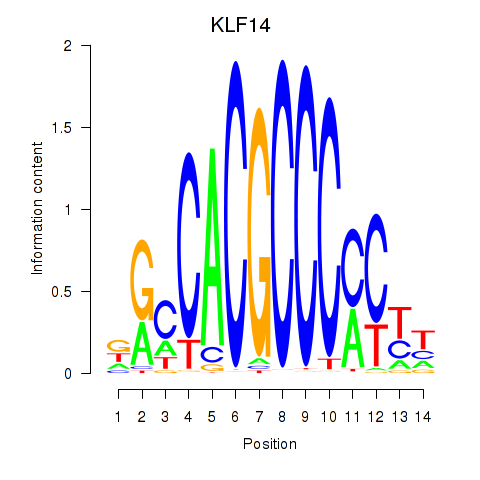
| KLF14_SP8 | -0.635 |
|
|
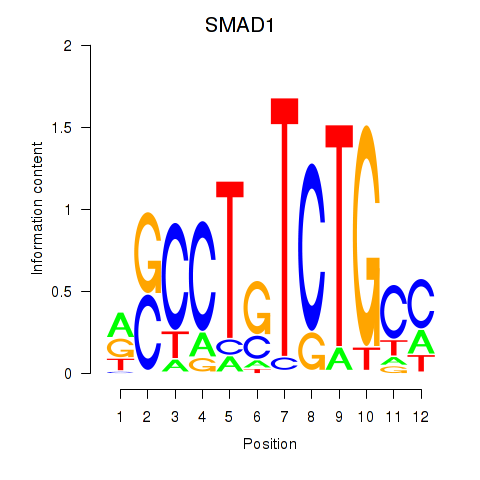
| SMAD1 | -0.638 |
|
|
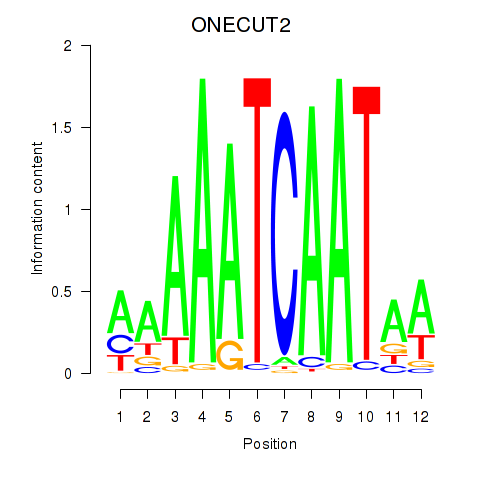
| ONECUT2_ONECUT3 | -0.638 |
|
|
| HOXC8 | -0.661 |
|
|
| IRF9 | -0.671 |
|
|
| THRB | -0.680 |
|
|
| ALX1_ARX | -0.683 |
|
|
| KLF12 | -0.696 |
|
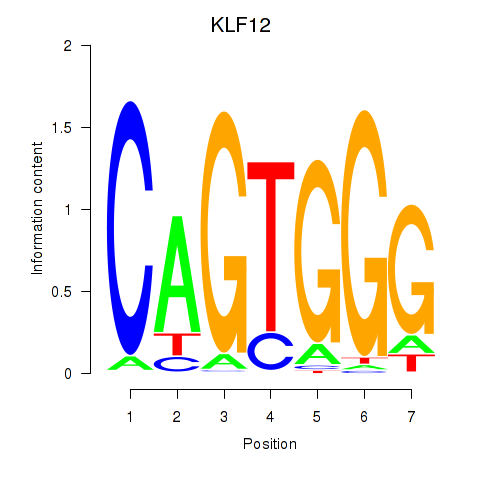
|
| HES1 | -0.709 |
|
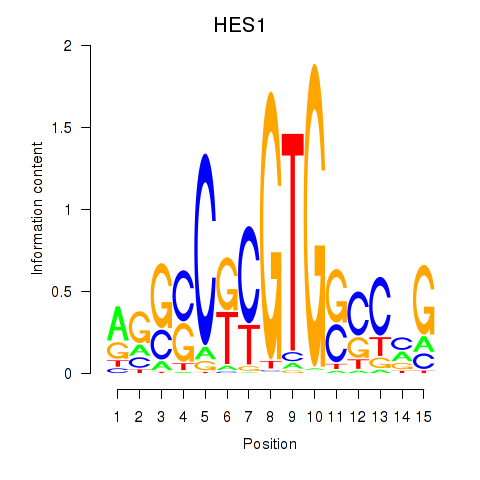
|
| THRA_RXRB | -0.712 |
|
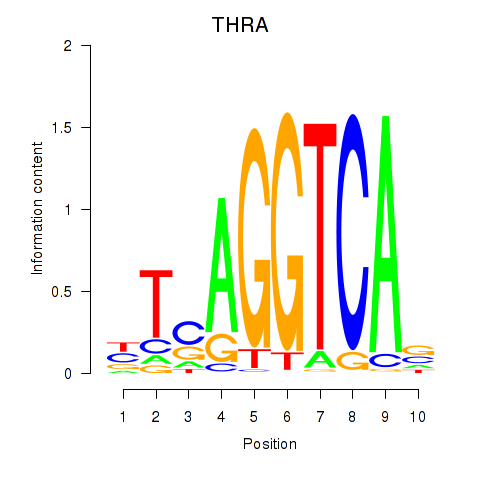
|
| HOXC6_HOXA7 | -0.730 |
|
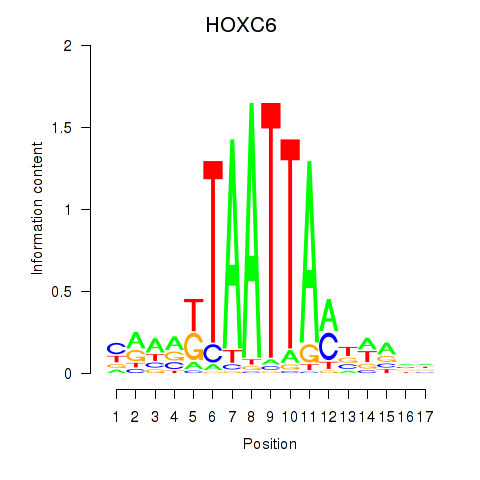
|
| EGR3_EGR2 | -0.734 |
|
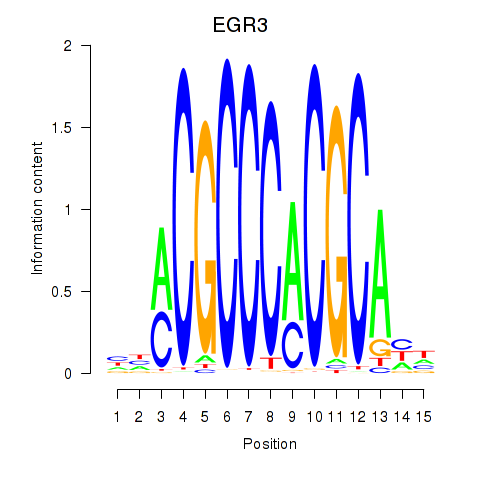
|
| SOX21 | -0.737 |
|
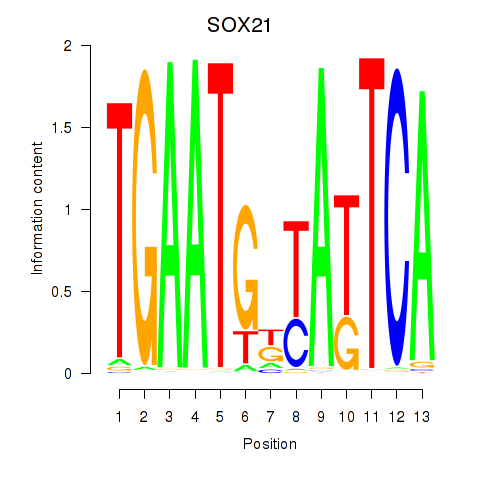
|
| LHX6 | -0.742 |
|
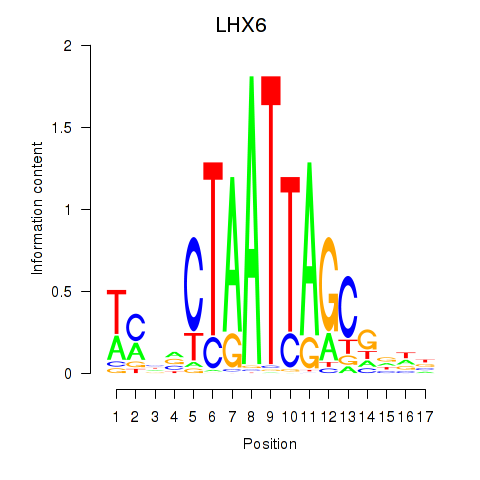
|
| MBD2 | -0.745 |
|
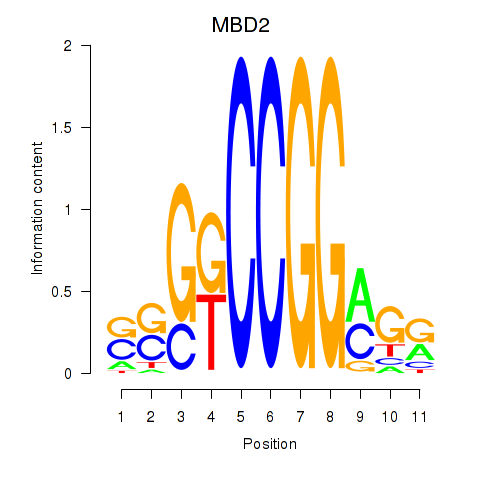
|
| ZIC3_ZIC4 | -0.761 |
|
|
| FOXC1 | -0.777 |
|
|
| ZIC2_GLI1 | -0.783 |
|
|
| FOXF2_FOXJ1 | -0.784 |
|
|
| ZNF8 | -0.784 |
|
|
| CENPB | -0.784 |
|
|
| SHOX2_HOXC5 | -0.785 |
|
|
| DLX3_EVX1_MEOX1 | -0.812 |
|
|
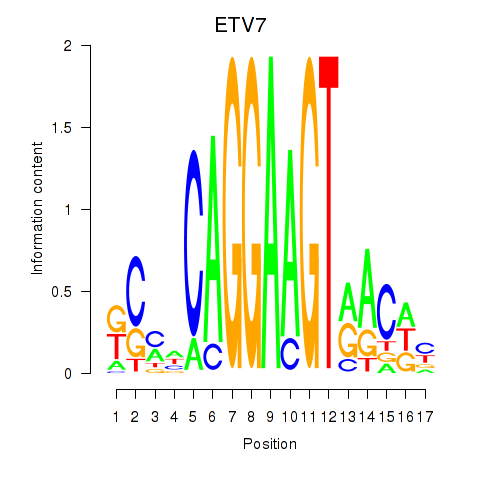
| ETV7 | -0.818 |
|
|
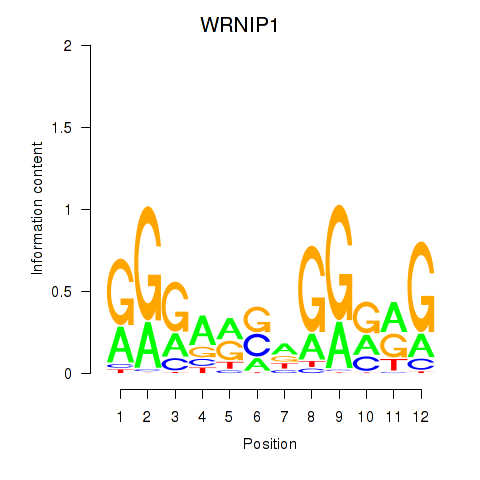
| WRNIP1 | -0.822 |
|
|
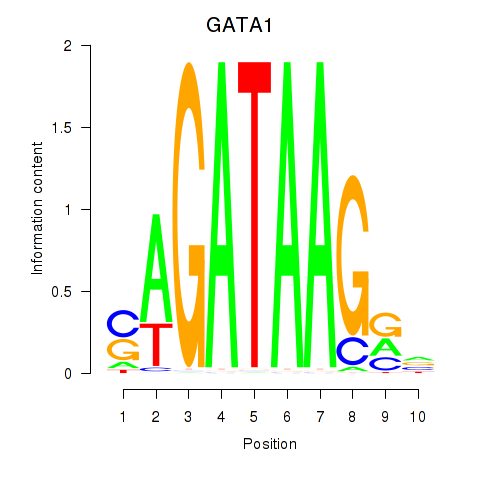
| GATA1_GATA4 | -0.826 |
|
|
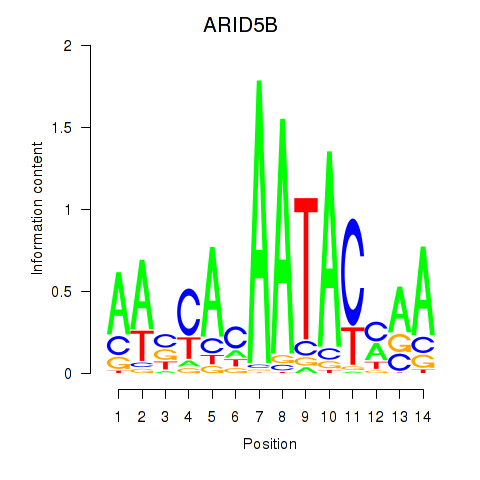
| ARID5B | -0.831 |
|
|
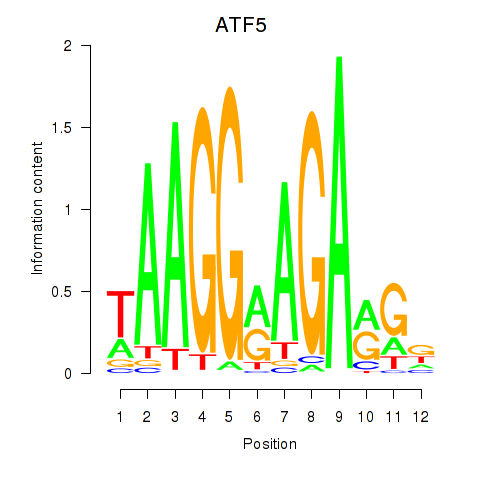
| ATF5 | -0.857 |
|
|
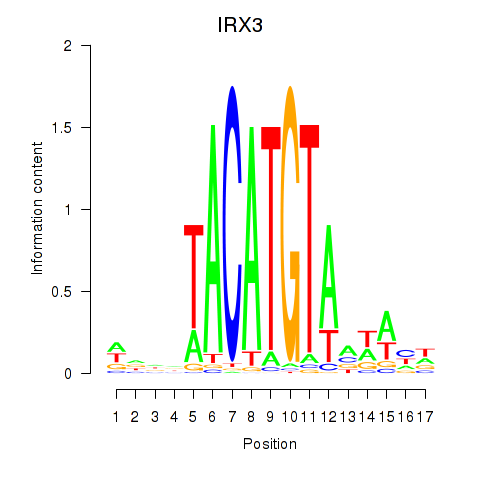
| IRX3 | -0.879 |
|
|
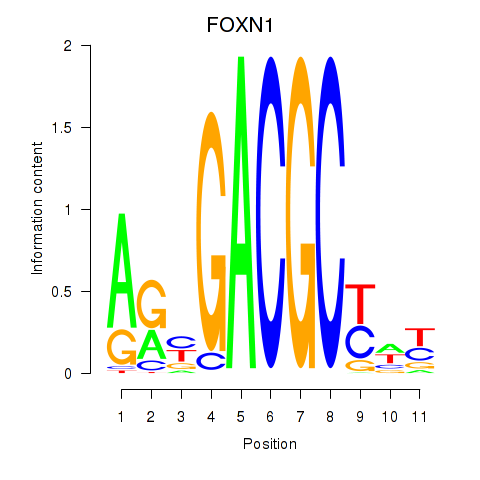
| FOXN1 | -0.880 |
|
|
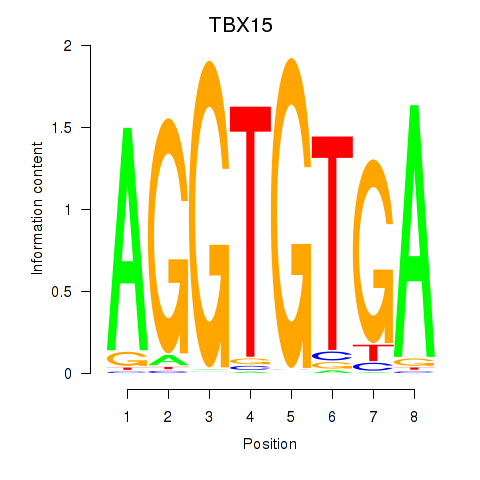
| TBX15_MGA | -0.881 |
|
|
| JUN | -0.883 |
|
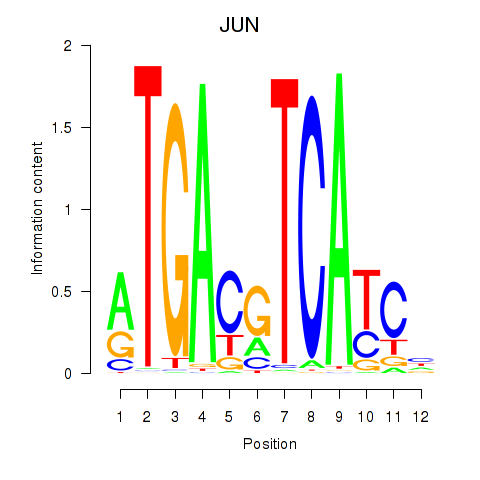
|
| MXI1_MYC_MYCN | -0.891 |
|
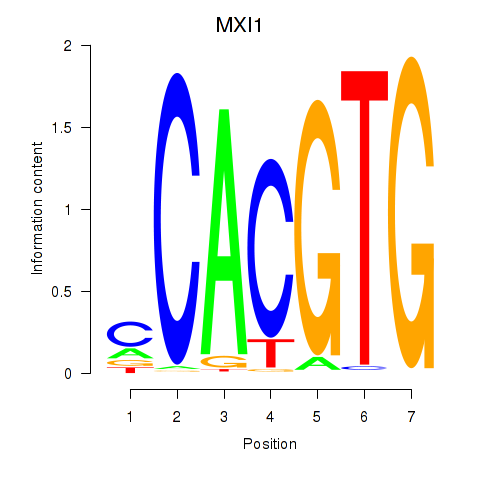
|
| ELF2_GABPA_ELF5 | -0.911 |
|
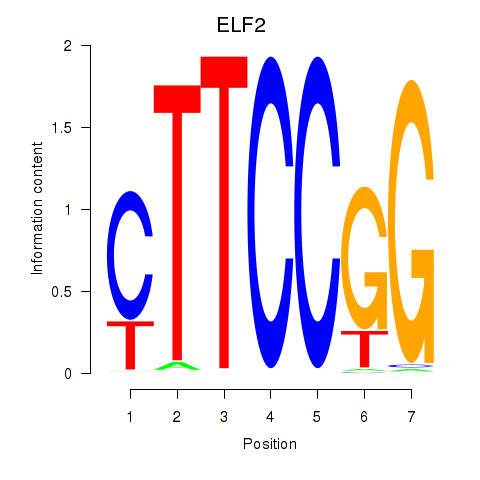
|
| CREB3L1_CREB3 | -0.918 |
|
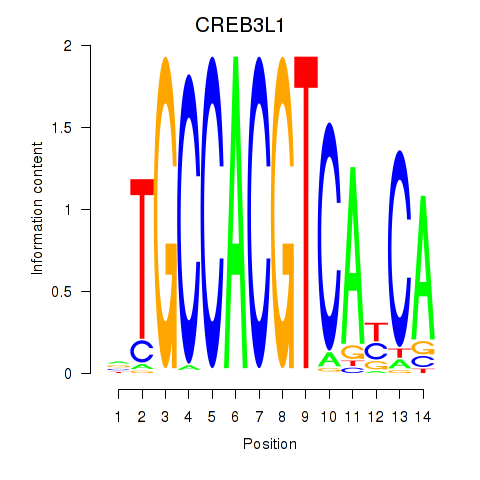
|
| ONECUT1 | -0.920 |
|
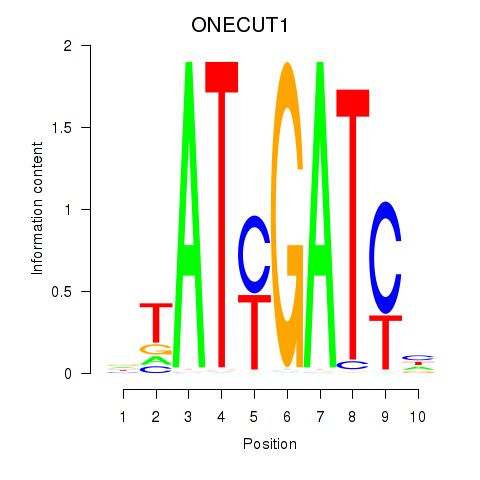
|
| SOX11 | -0.923 |
|
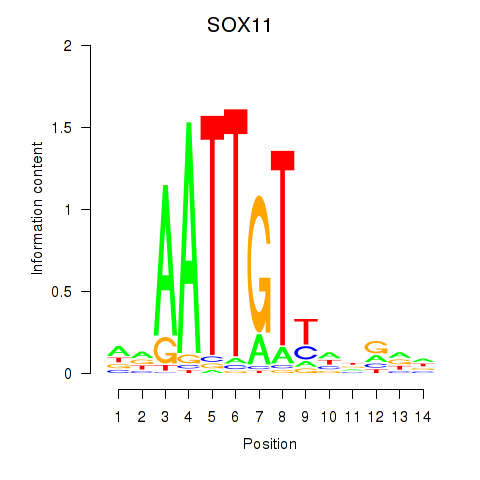
|
| PRDM14 | -0.949 |
|
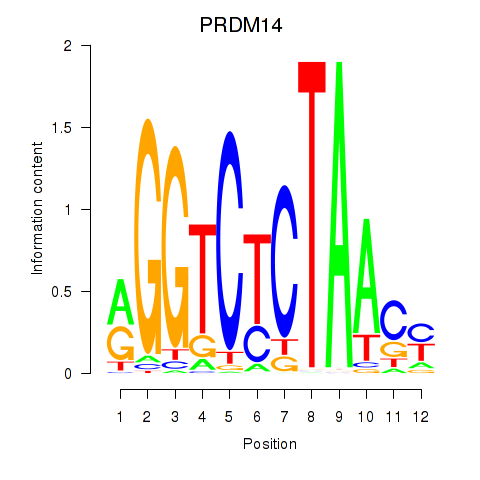
|
| GLI2 | -0.951 |
|
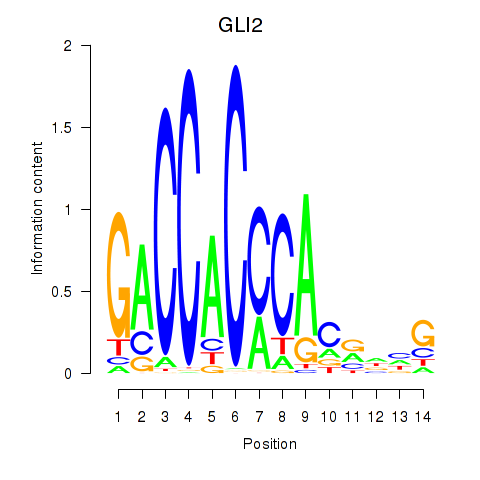
|
| HDX | -0.978 |
|
|
| CUX1 | -1.009 |
|
|
| NR0B1 | -1.015 |
|
|
| PAX1_PAX9 | -1.046 |
|
|
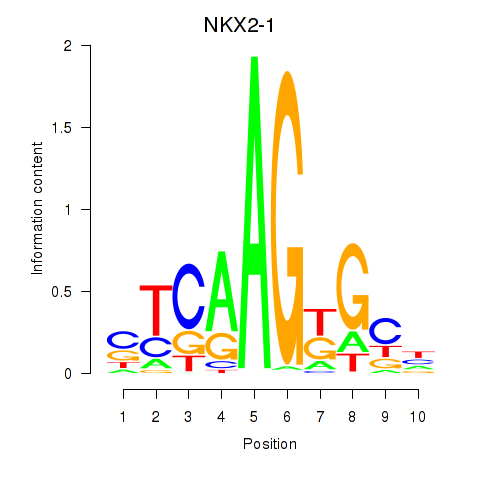
| NKX2-1 | -1.056 |
|
|
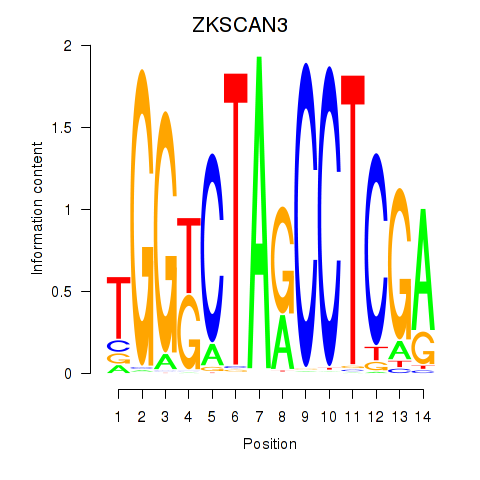
| ZKSCAN3 | -1.059 |
|
|
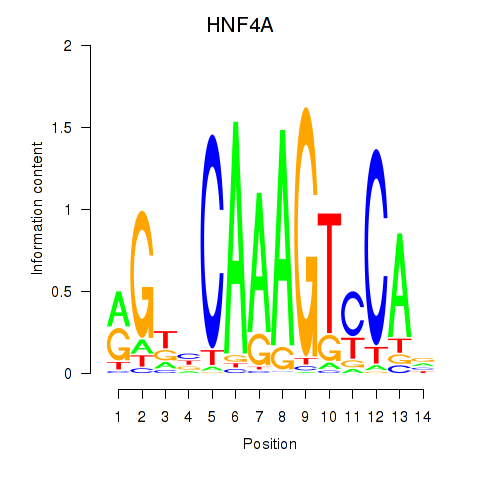
| HNF4A | -1.067 |
|
|
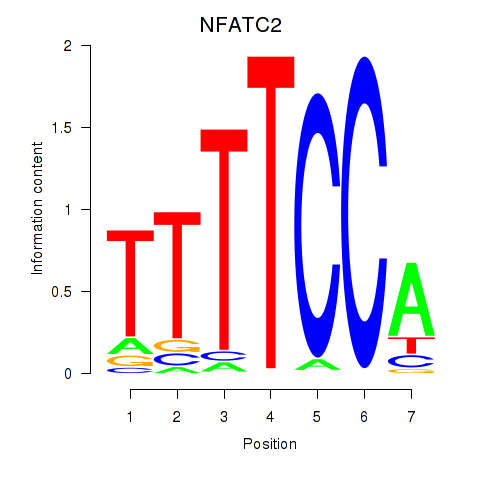
| NFATC2_NFATC3 | -1.094 |
|
|
| SPIB | -1.126 |
|
|
| NR1D1 | -1.143 |
|
|
| ZSCAN4 | -1.149 |
|
|
| FOXO3_FOXD2 | -1.150 |
|
|
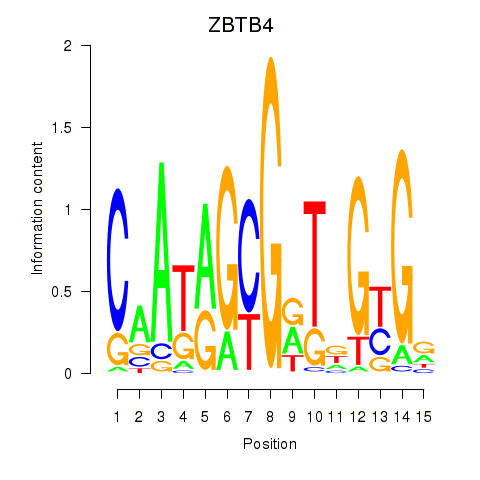
| ZBTB4 | -1.152 |
|
|
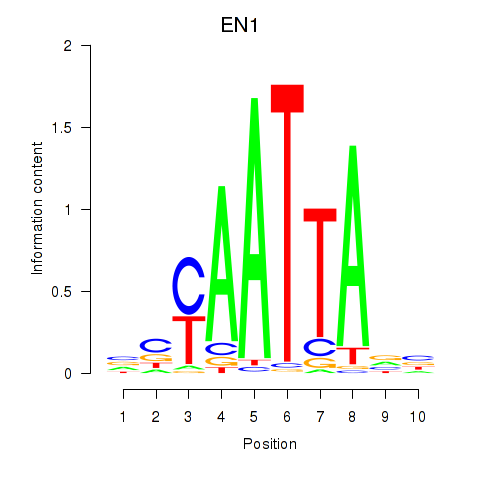
| EN1_ESX1_GBX1 | -1.168 |
|
|
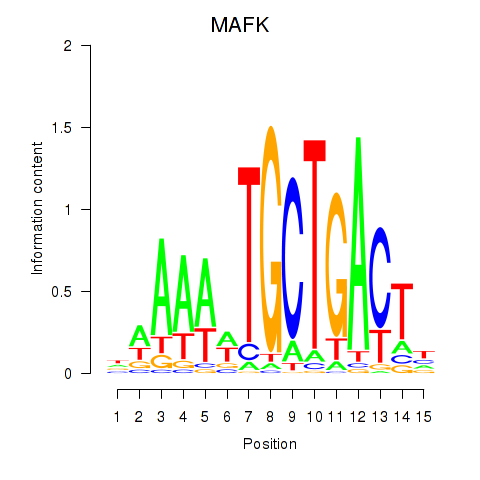
| MAFK | -1.189 |
|
|
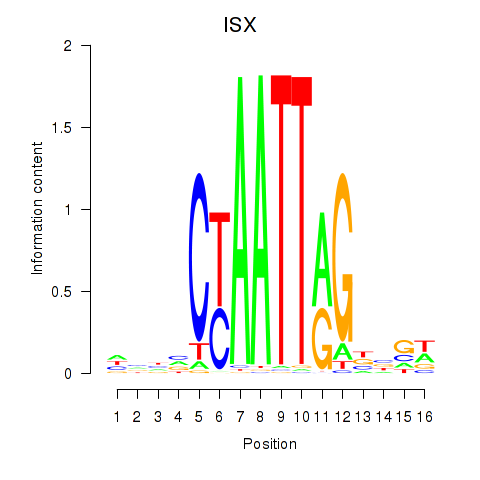
| ISX | -1.199 |
|
|
| KLF16_SP2 | -1.232 |
|
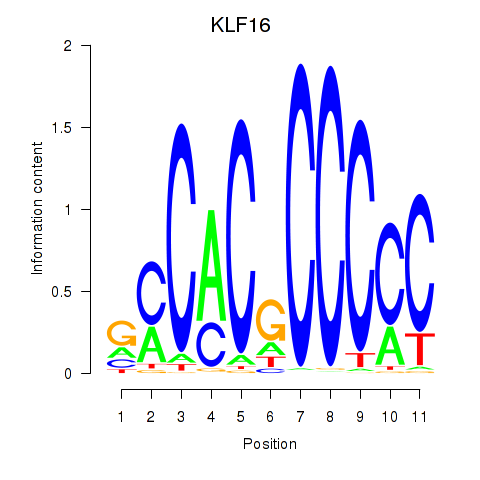
|
| NR1H4 | -1.242 |
|
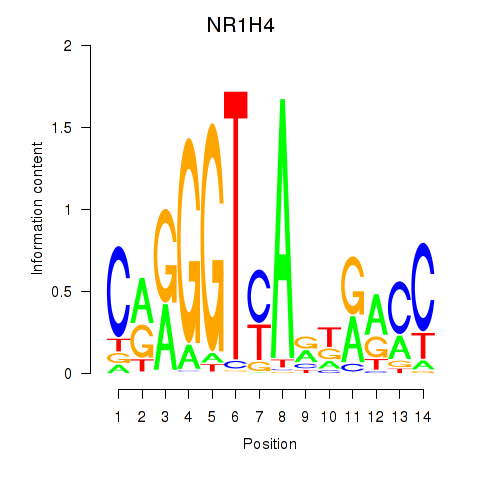
|
| MEF2D_MEF2A | -1.251 |
|
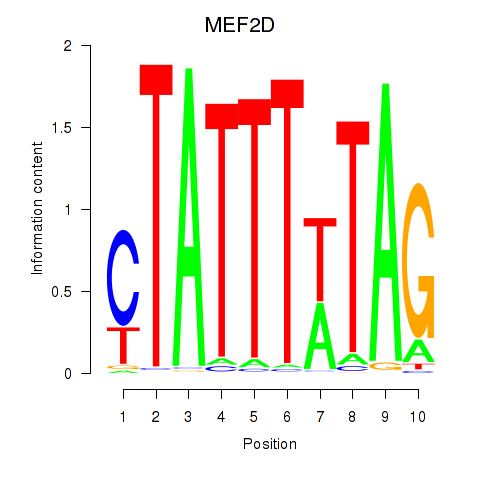
|
| SOX3_SOX2 | -1.258 |
|
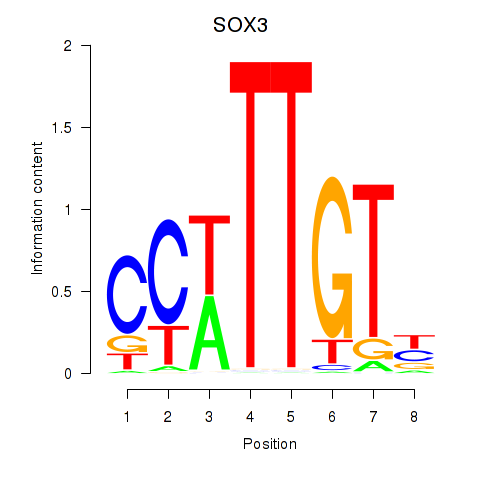
|
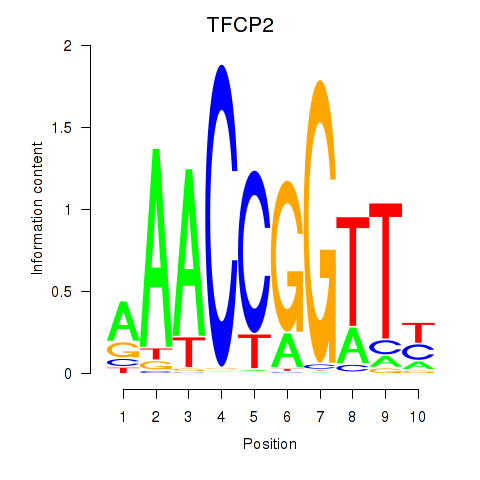
| TFCP2 | -1.262 |
|
|
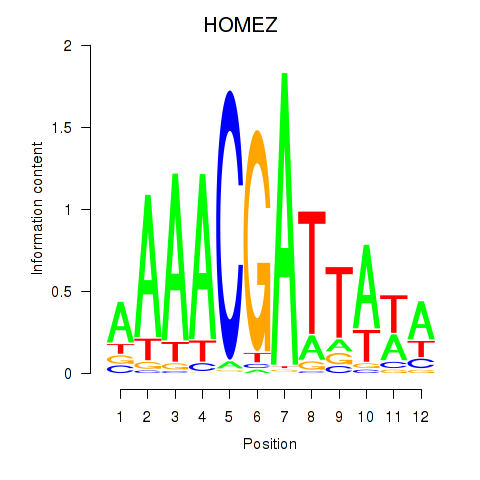
| HOMEZ | -1.270 |
|
|
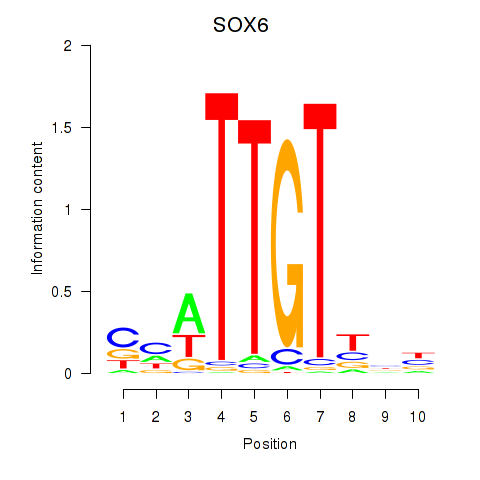
| SOX6 | -1.274 |
|
|
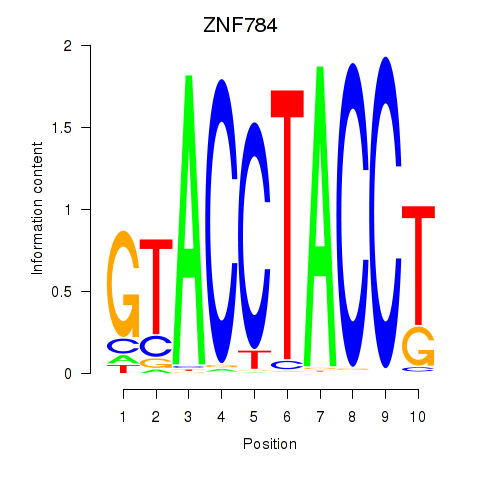
| ZNF784 | -1.275 |
|
|
| LHX2 | -1.289 |
|
|
| RXRA_NR2F6_NR2C2 | -1.295 |
|
|
| GFI1 | -1.318 |
|
|
| GCM2 | -1.328 |
|
|
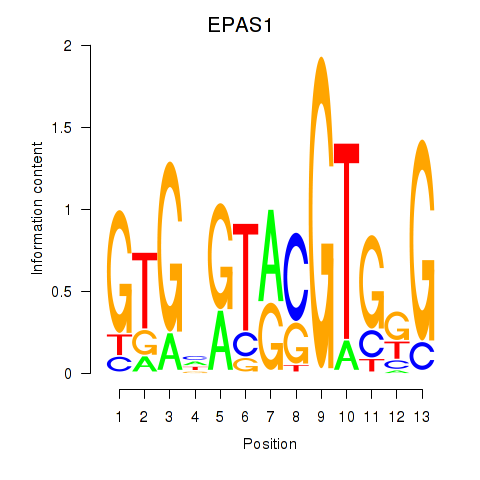
| EPAS1_BCL3 | -1.331 |
|
|
| NR5A2 | -1.346 |
|
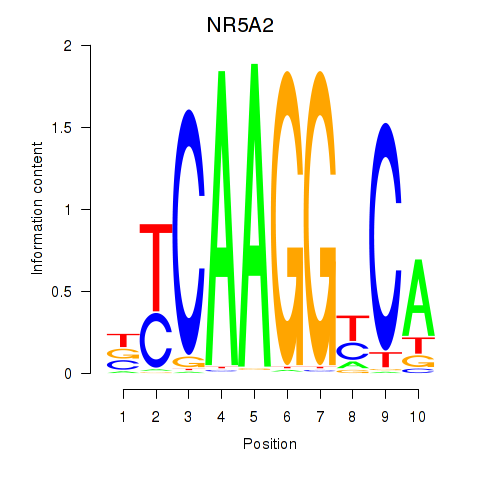
|
| MYB | -1.355 |
|
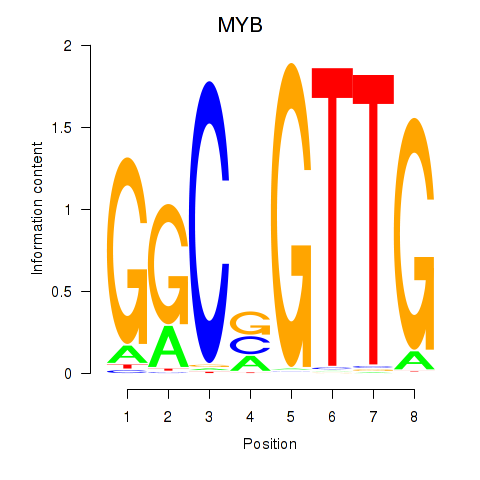
|
| SMAD3 | -1.367 |
|
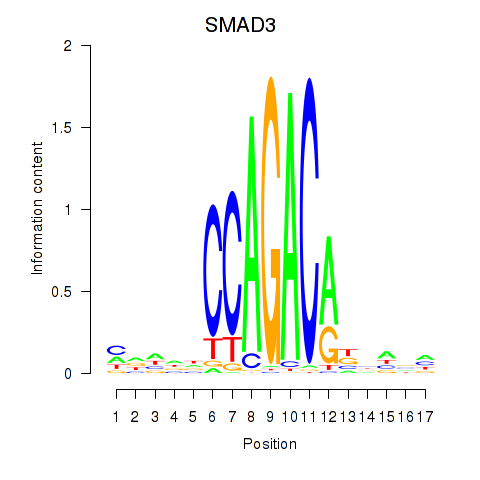
|
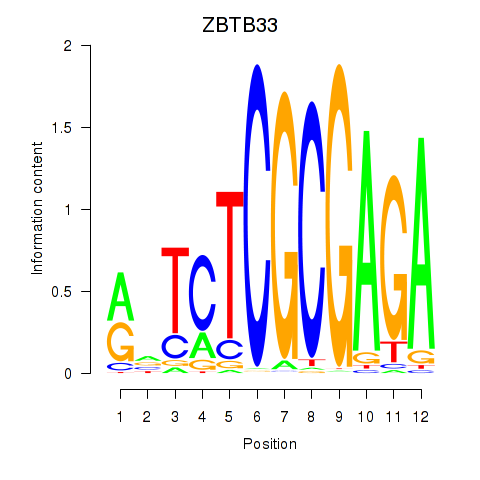
| ZBTB33_CHD2 | -1.392 |
|
|
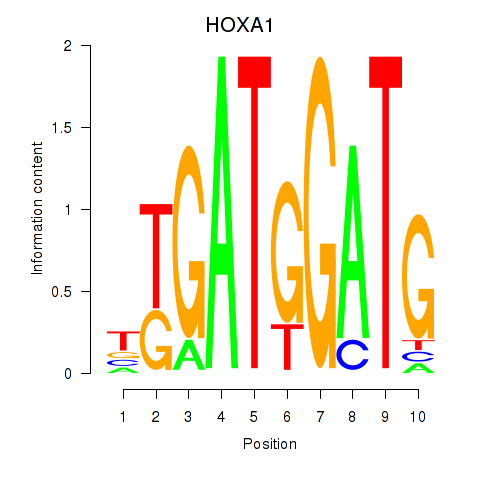
| HOXA1 | -1.405 |
|
|
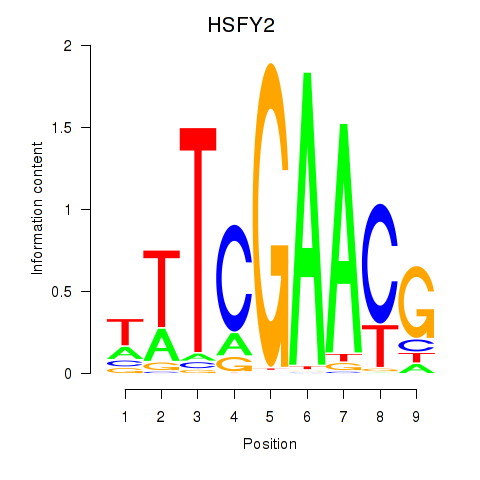
| HSFY2 | -1.411 |
|
|
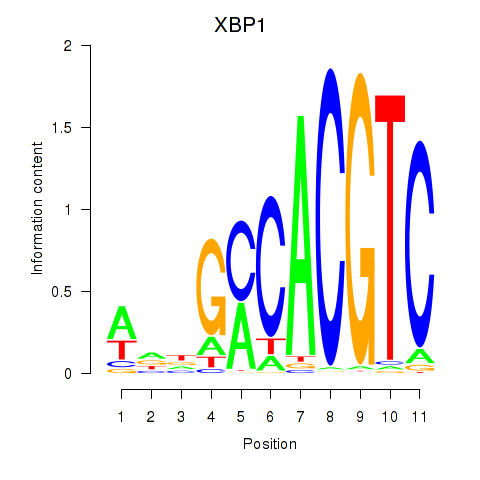
| XBP1 | -1.429 |
|
|
| DDIT3 | -1.438 |
|
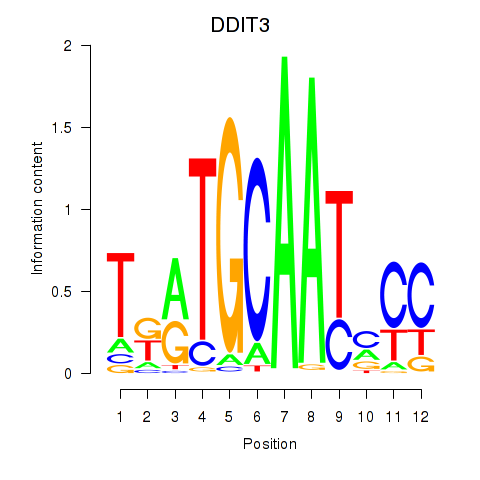
|
| PRDM1 | -1.452 |
|
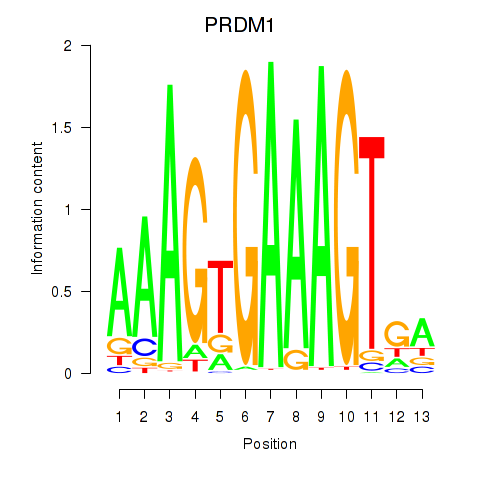
|
| PBX3 | -1.455 |
|
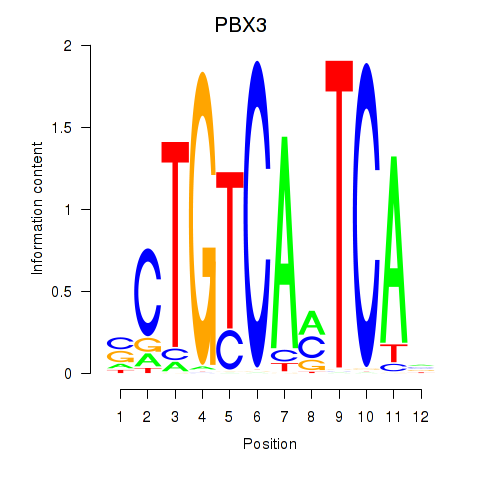
|
| SOX8 | -1.458 |
|
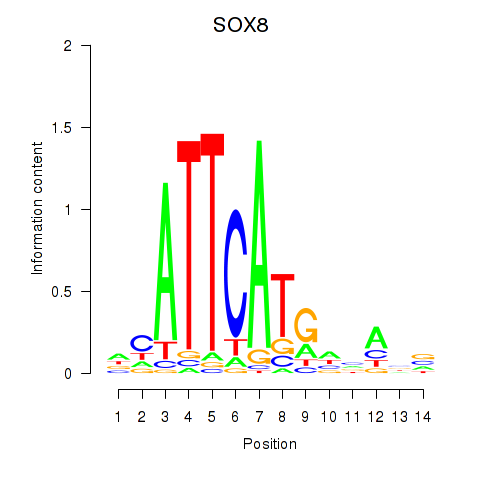
|
| HMX2 | -1.485 |
|
|
| PRDM4 | -1.539 |
|
|
| TLX2 | -1.569 |
|
|
| OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 | -1.575 |
|
|
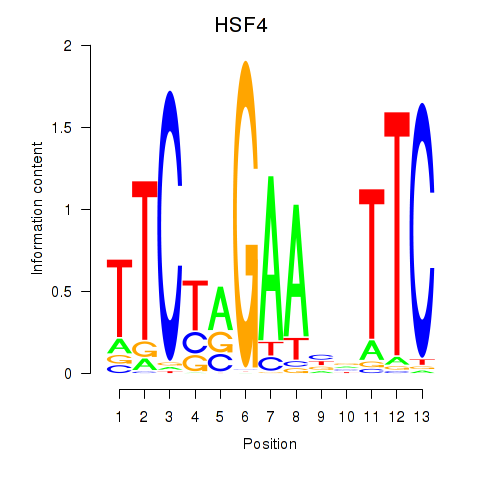
| HSF4 | -1.578 |
|
|
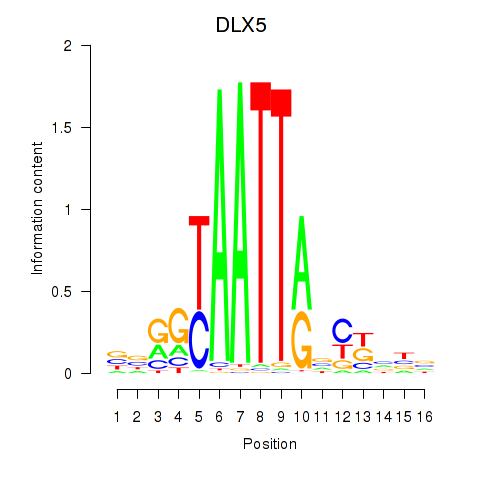
| DLX5 | -1.579 |
|
|
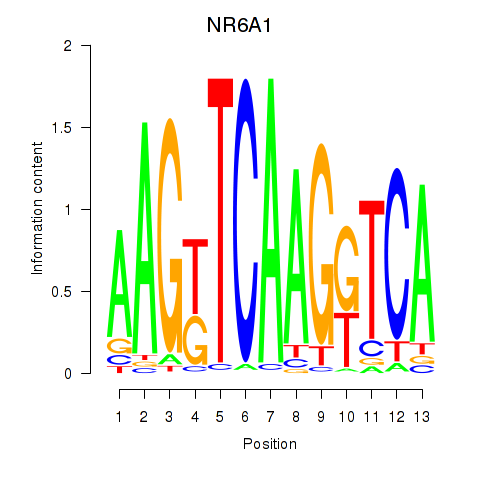
| NR6A1 | -1.642 |
|
|
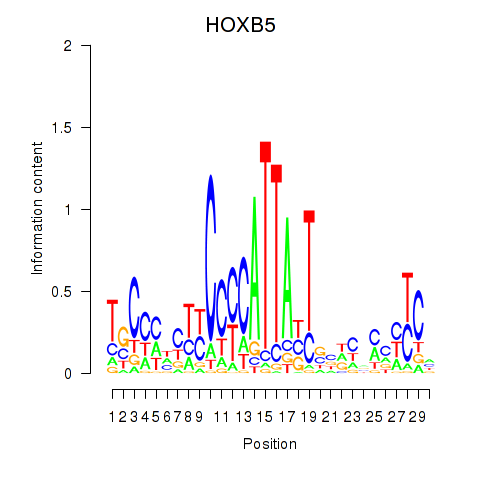
| HOXB5 | -1.655 |
|
|
| PAX8 | -1.685 |
|
|
| MESP1 | -1.744 |
|
|
| IKZF1 | -1.745 |
|
|
| HOXC12_HOXD12 | -1.758 |
|
|
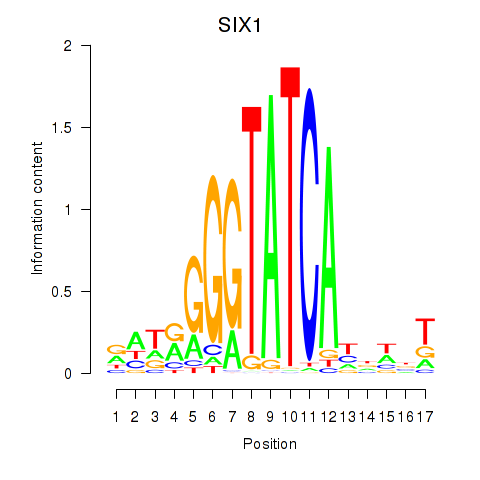
| SIX1_SIX3_SIX2 | -1.784 |
|
|
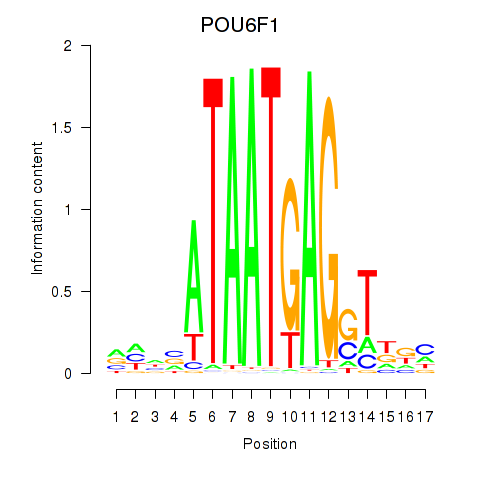
| POU6F1 | -1.814 |
|
|
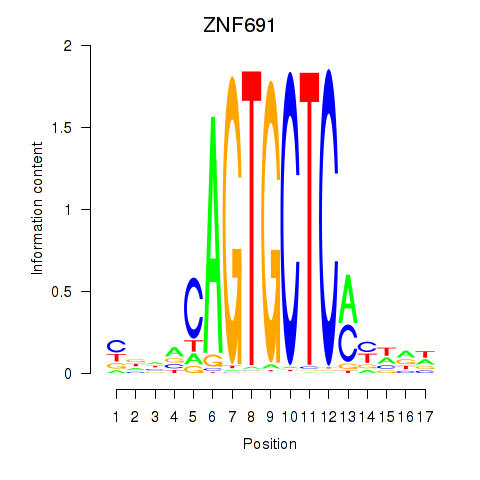
| ZNF691 | -1.823 |
|
|
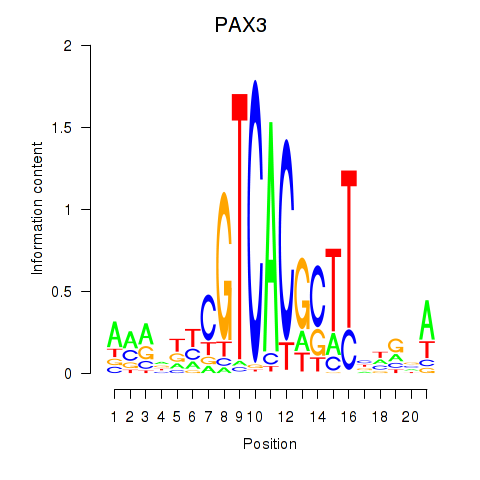
| PAX3 | -1.825 |
|
|
| NKX2-5 | -1.832 |
|
|
| FOXJ2 | -1.834 |
|
|
| TCF21 | -1.853 |
|
|
| CTCF_CTCFL | -1.854 |
|
|
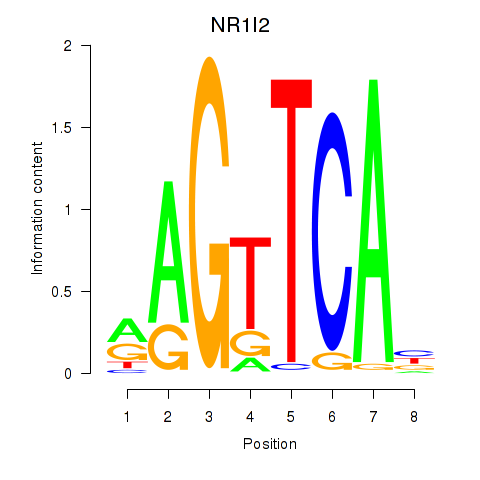
| NR1I2 | -1.869 |
|
|
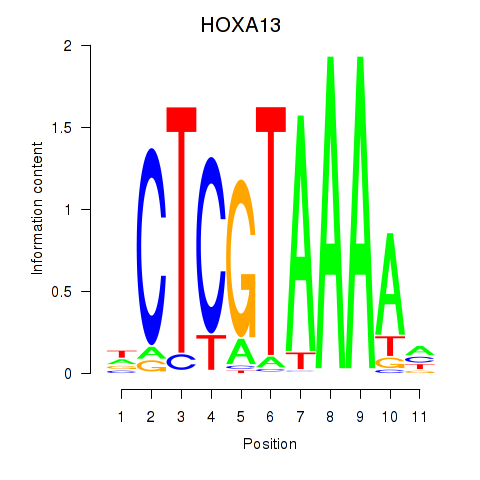
| HOXA13 | -1.895 |
|
|
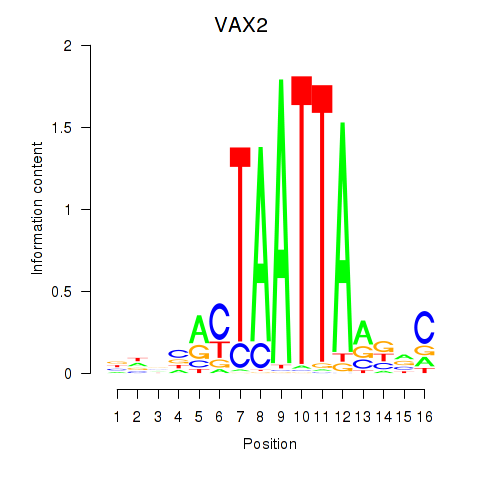
| VAX2_RHOXF2 | -1.896 |
|
|
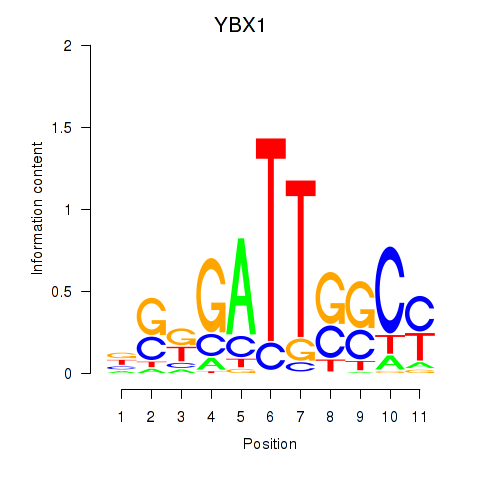
| YBX1_FOS_NFYC_NFYA_NFYB_CEBPZ | -1.978 |
|
|
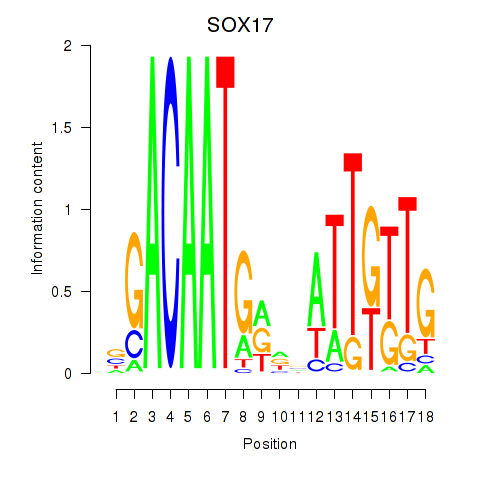
| SOX17 | -1.986 |
|
|
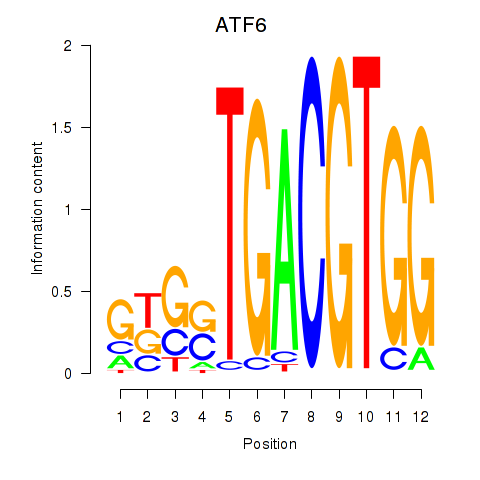
| ATF6 | -1.986 |
|
|
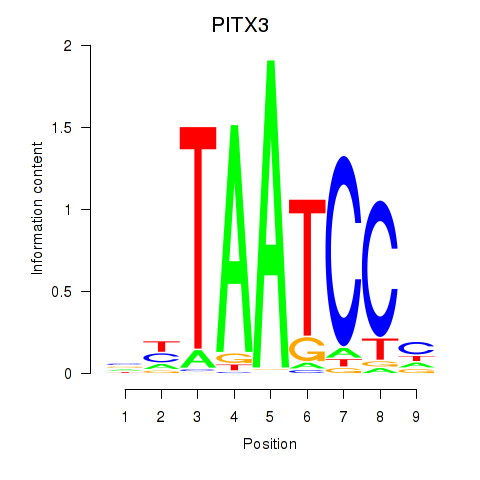
| PITX3 | -2.045 |
|
|
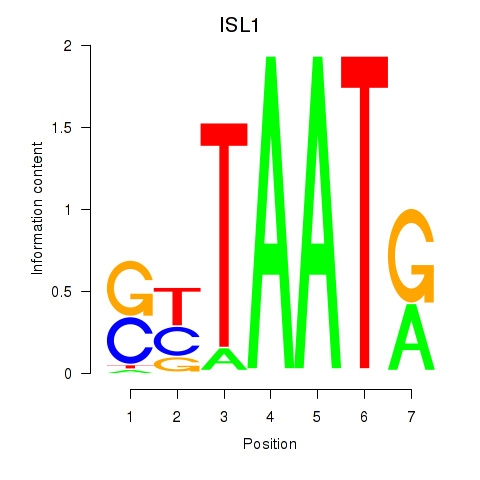
| ISL1 | -2.055 |
|
|
| MAX_TFEB | -2.055 |
|
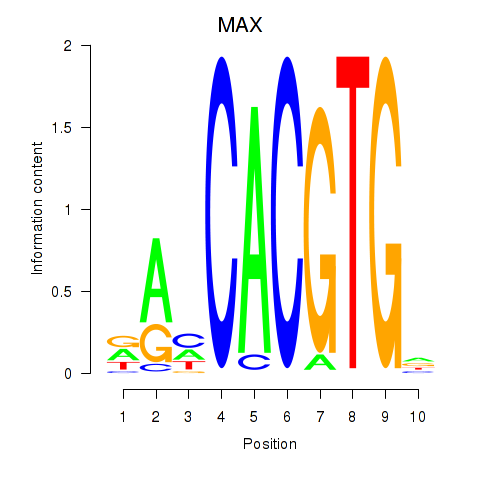
|
| RELB | -2.117 |
|
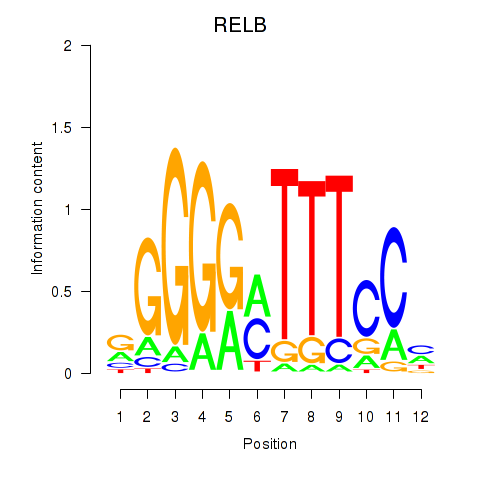
|
| RARG | -2.182 |
|
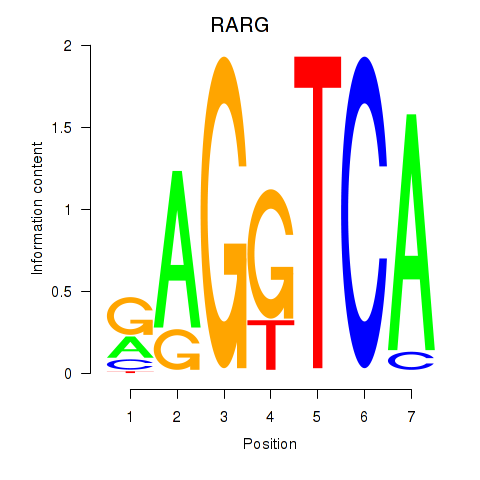
|
| MZF1 | -2.208 |
|
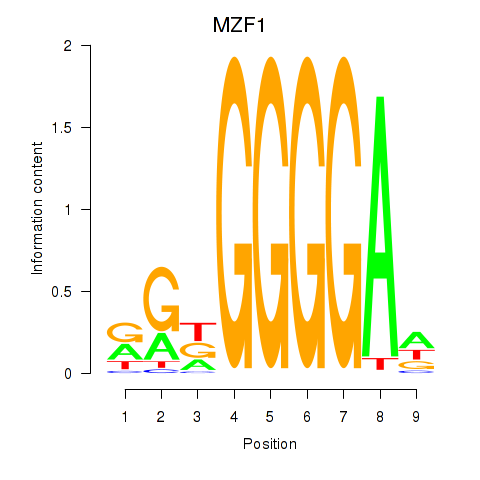
|
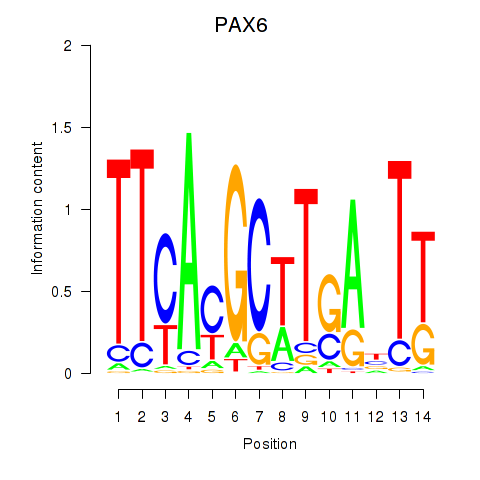
| PAX6 | -2.212 |
|
|
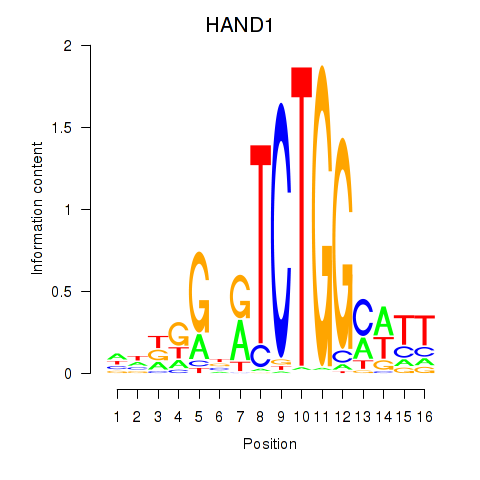
| HAND1 | -2.252 |
|
|
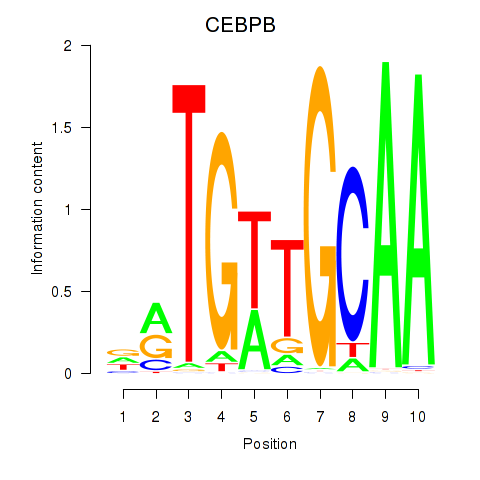
| CEBPB | -2.256 |
|
|
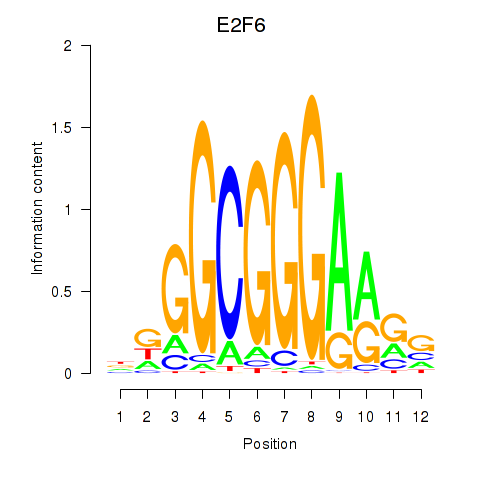
| E2F6 | -2.271 |
|
|
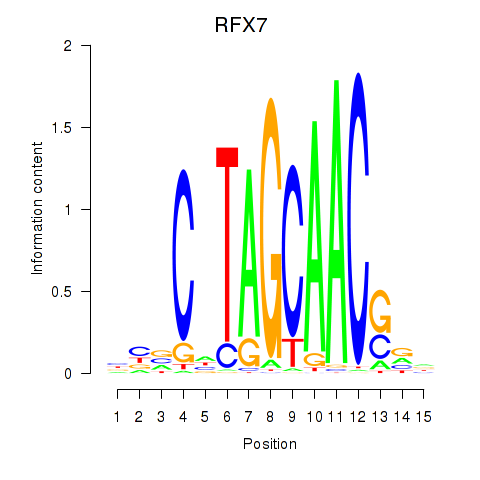
| RFX7_RFX4_RFX1 | -2.283 |
|
|
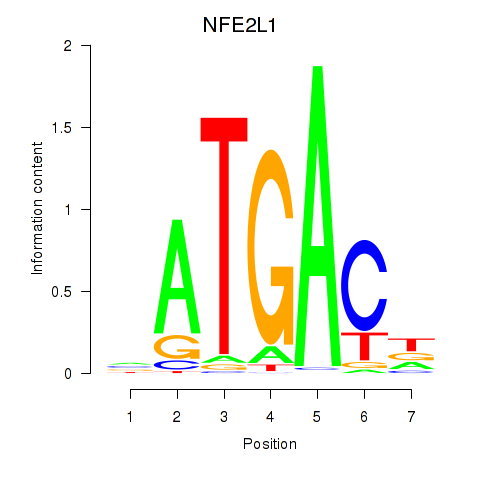
| NFE2L1 | -2.287 |
|
|
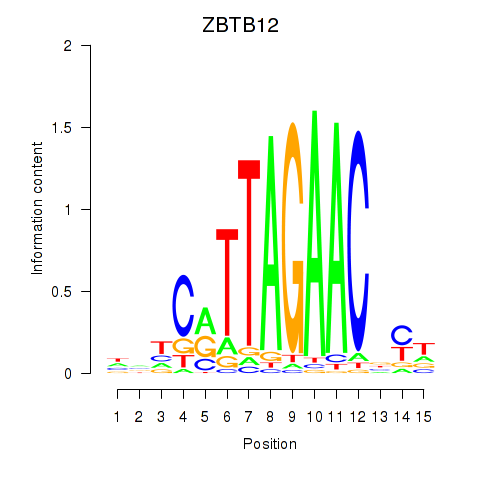
| ZBTB12 | -2.306 |
|
|
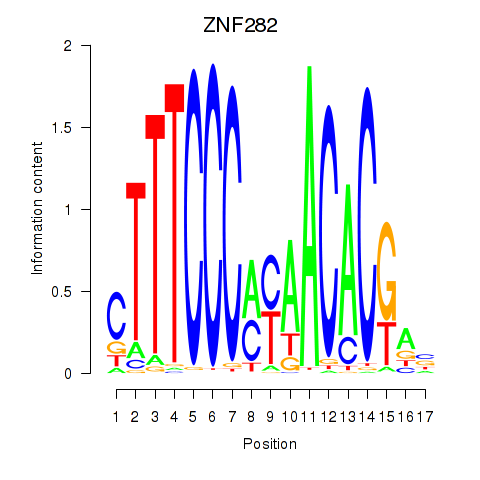
| ZNF282 | -2.318 |
|
|
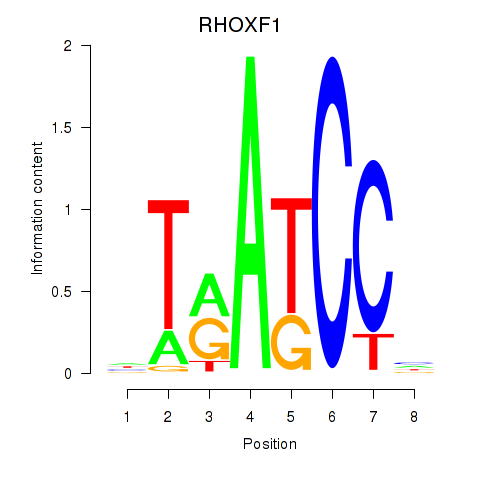
| RHOXF1 | -2.432 |
|
|
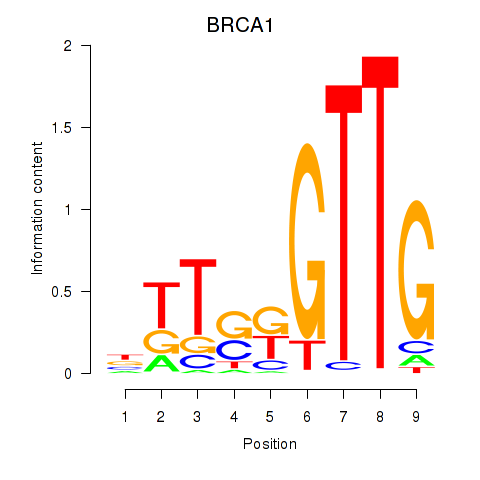
| BRCA1 | -2.446 |
|
|
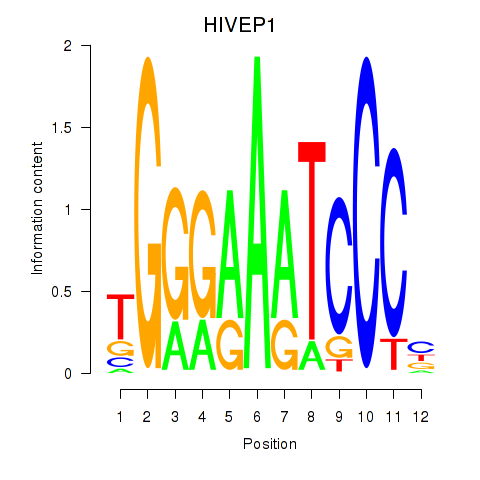
| HIVEP1 | -2.473 |
|
|
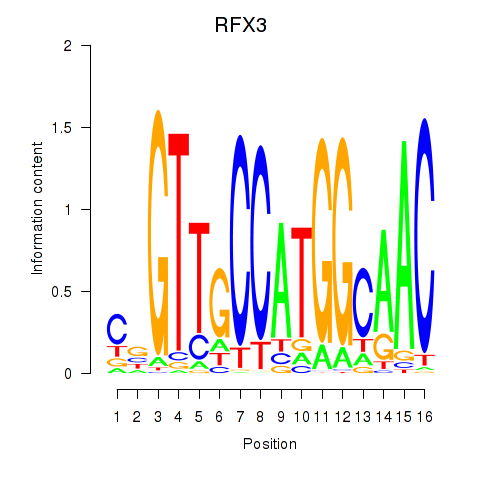
| RFX3_RFX2 | -2.480 |
|
|
| RARA | -2.498 |
|
|
| TEAD4 | -2.507 |
|
|
| ZNF274 | -2.538 |
|
|
| SOX18 | -2.616 |
|
|
| BATF | -2.645 |
|
|
| ZBTB3 | -2.654 |
|
|
| CXXC1 | -2.665 |
|
|
| AIRE | -2.679 |
|
|
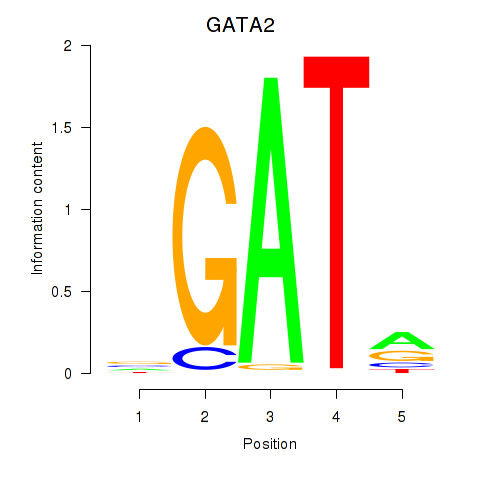
| GATA2 | -2.711 |
|
|
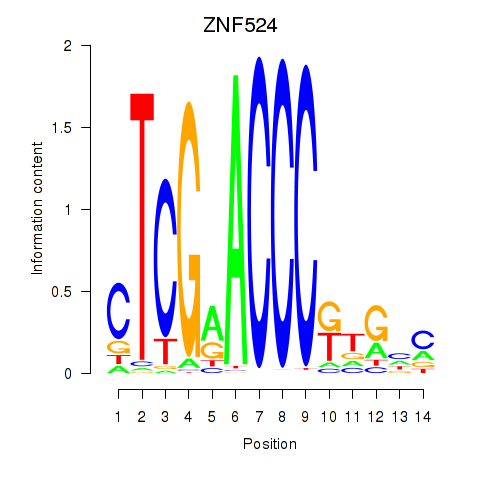
| ZNF524 | -2.750 |
|
|
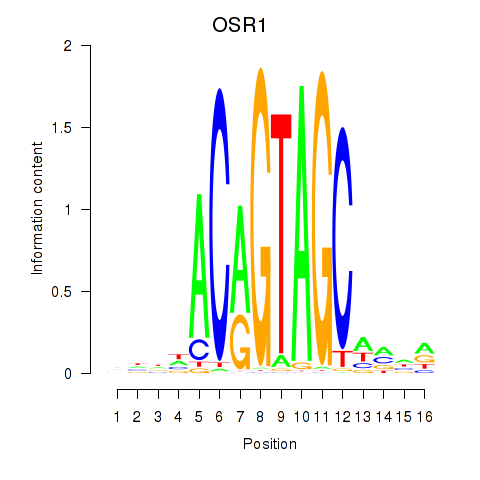
| OSR1_OSR2 | -2.758 |
|
|
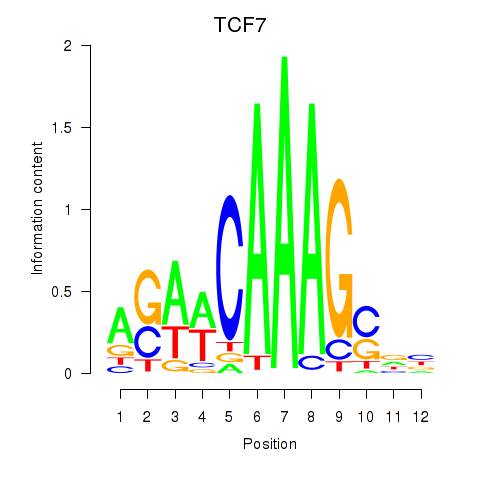
| TCF7 | -2.762 |
|
|
| MAFF_MAFG | -2.790 |
|
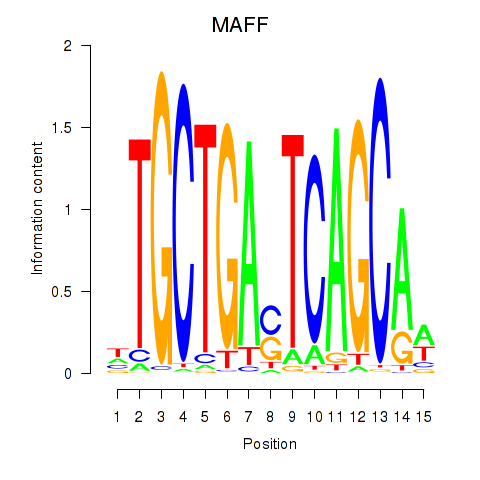
|
| ZNF384 | -2.801 |
|
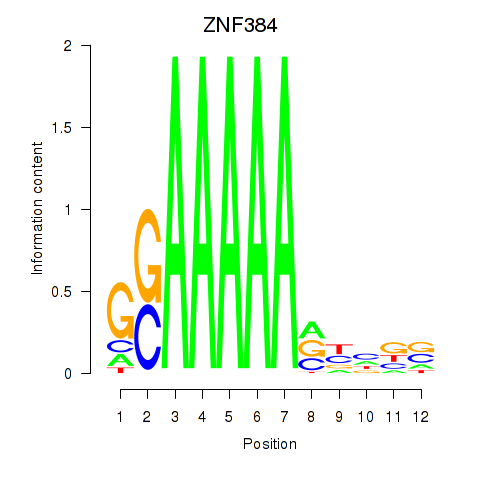
|
| NKX3-2 | -2.816 |
|
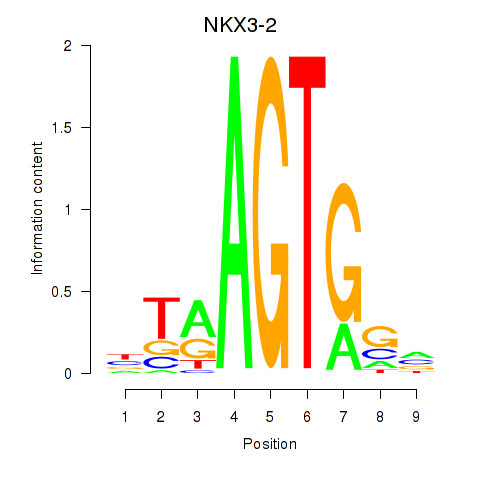
|
| SREBF1_TFE3 | -2.856 |
|
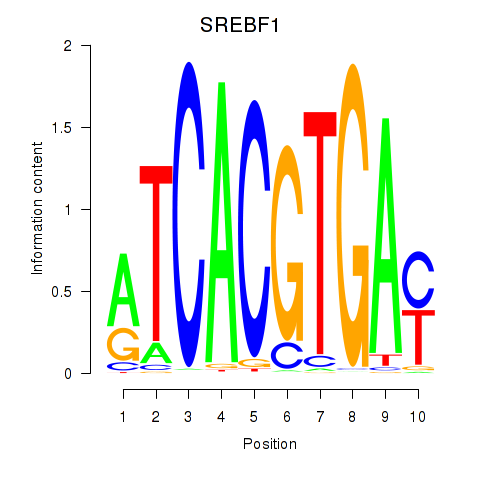
|
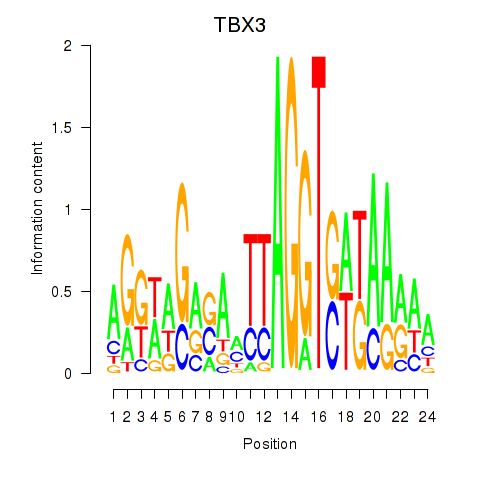
| TBX3 | -2.927 |
|
|
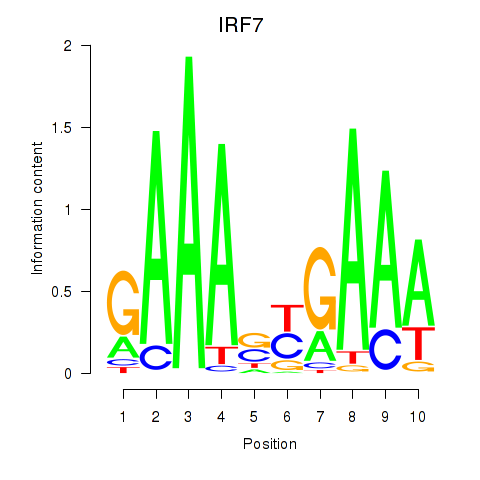
| IRF7 | -2.935 |
|
|
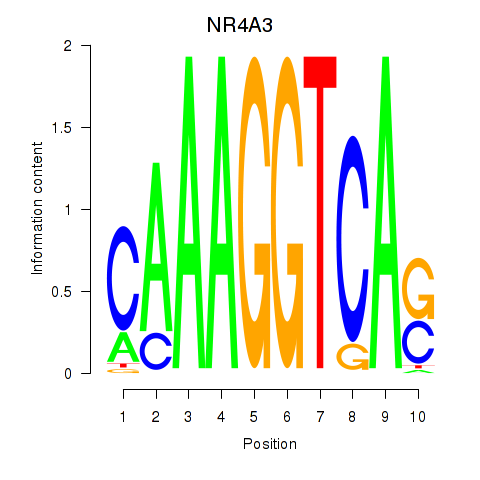
| NR4A3 | -3.086 |
|
|
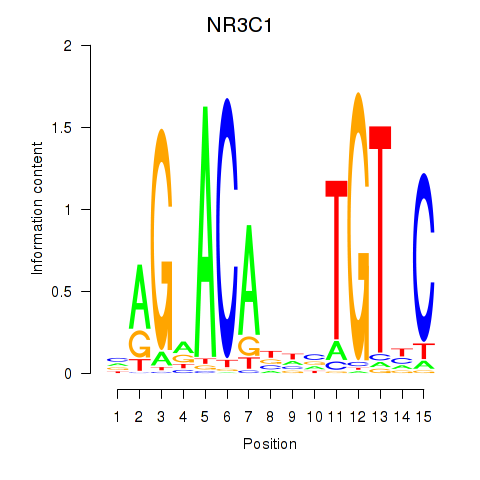
| NR3C1 | -3.197 |
|
|
| PAX4 | -3.212 |
|
|
| MAF_NRL | -3.226 |
|
|
| FOXK1_FOXP2_FOXB1_FOXP3 | -3.261 |
|
|
| TBX1 | -3.280 |
|
|
| IRX2 | -3.288 |
|
|
| NKX2-8 | -3.345 |
|
|
| TAF1 | -3.438 |
|
|
| PROX1 | -3.479 |
|
|
| OLIG3_NEUROD2_NEUROG2 | -3.504 |
|
|
| NRF1 | -3.583 |
|
|
| UBP1 | -3.602 |
|
|
| PITX1 | -3.667 |
|
|
| ZFX | -3.683 |
|
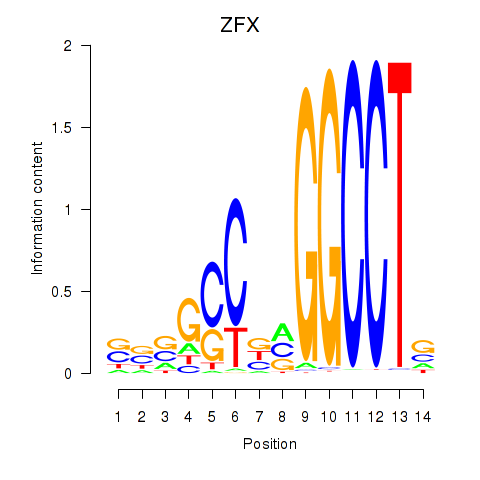
|
| BARHL1 | -3.757 |
|
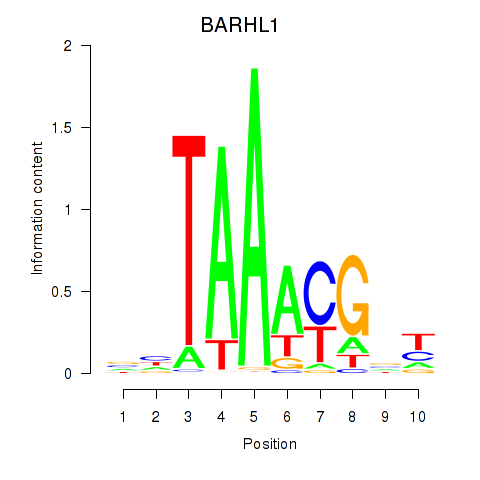
|
| POU5F1_POU2F3 | -3.773 |
|
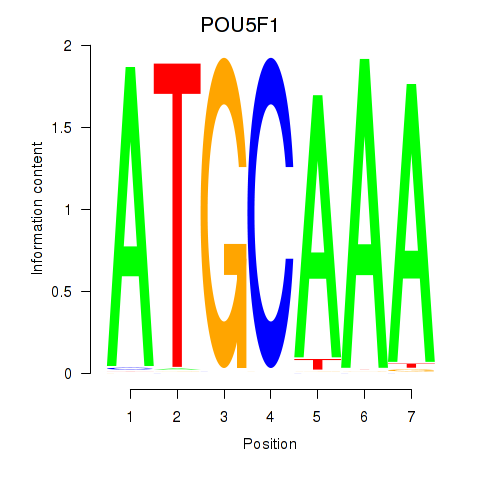
|
| YY1_YY2 | -3.817 |
|
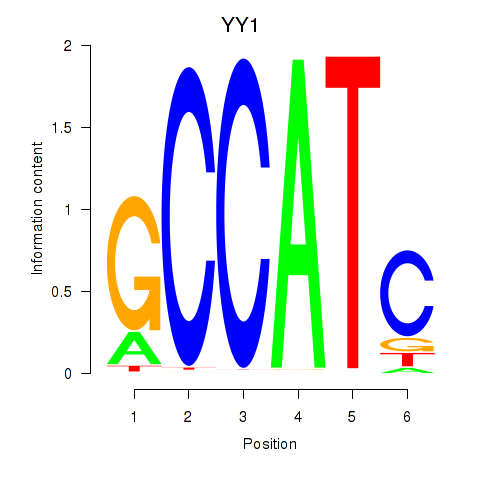
|
| SP100 | -3.830 |
|
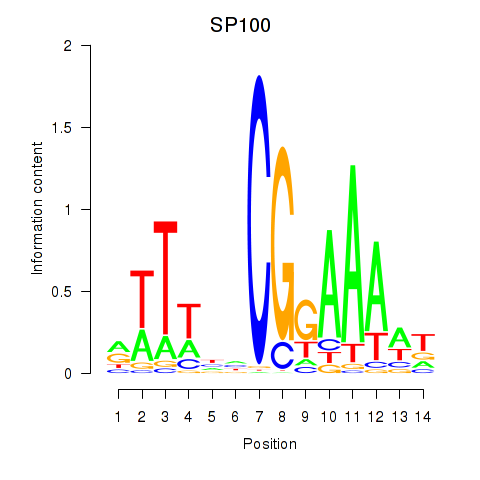
|
| IRF6_IRF4_IRF5 | -3.922 |
|
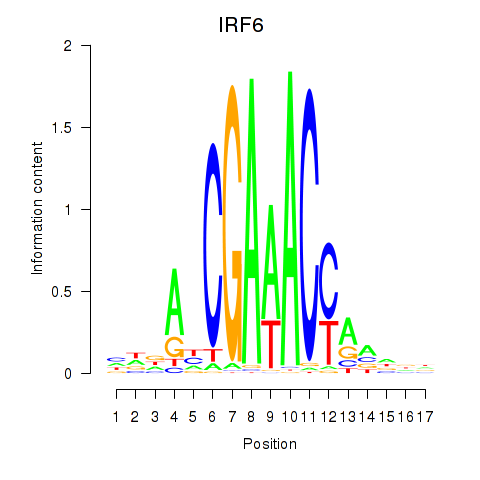
|
| NR2E3 | -3.936 |
|
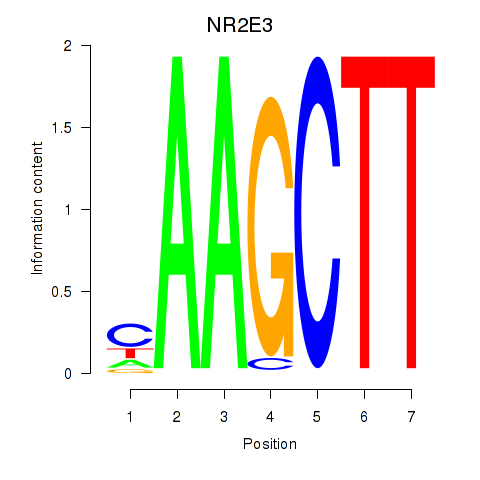
|
| IRF2_STAT2_IRF8_IRF1 | -4.086 |
|
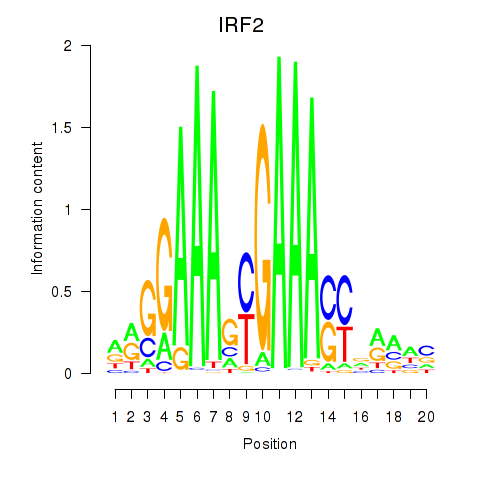
|
| ERG | -4.139 |
|
|
| BPTF | -4.271 |
|
|
| HOXA5 | -4.330 |
|
|
| PAX5 | -4.430 |
|
|
| SP1 | -4.732 |
|
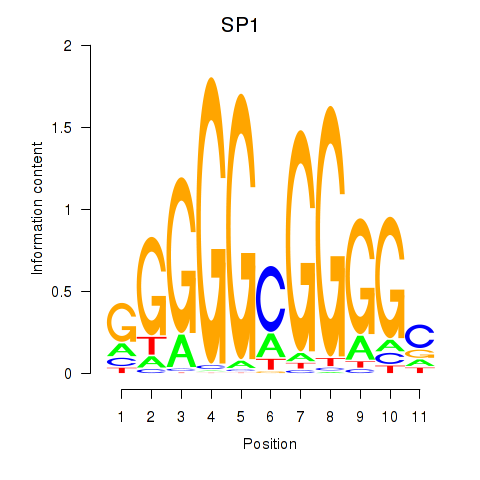
|
| ATF2_ATF1_ATF3 | -4.880 |
|
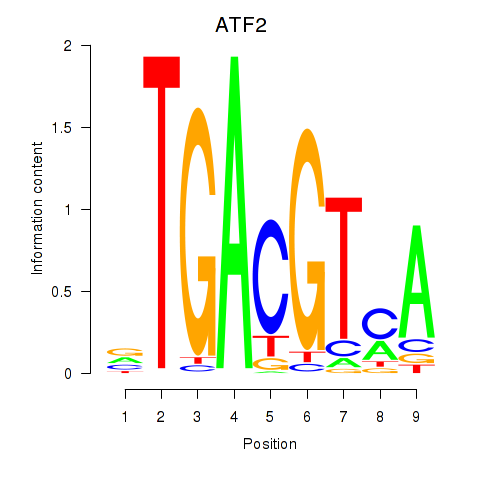
|
| HMBOX1 | -5.011 |
|
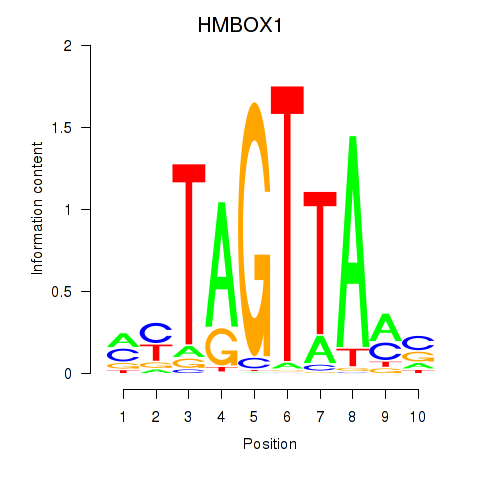
|
| MEIS2 | -5.026 |
|
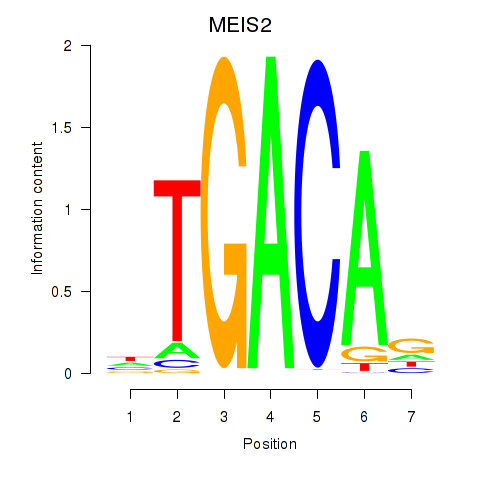
|
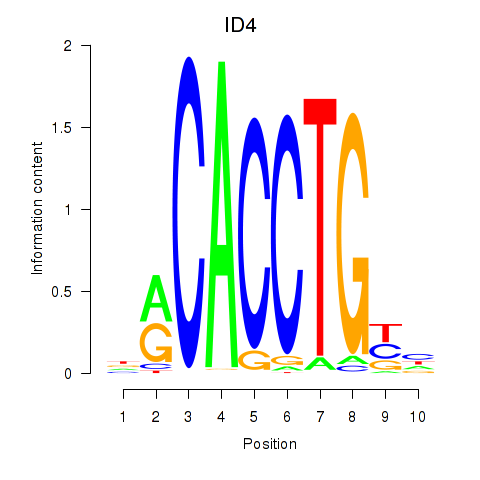
| ID4_TCF4_SNAI2 | -5.538 |
|
|
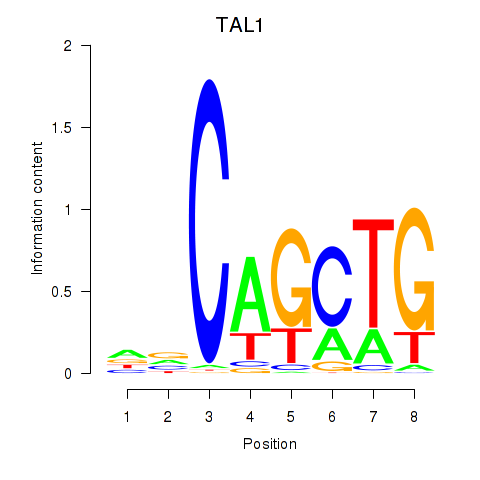
| TAL1 | -5.622 |
|
|
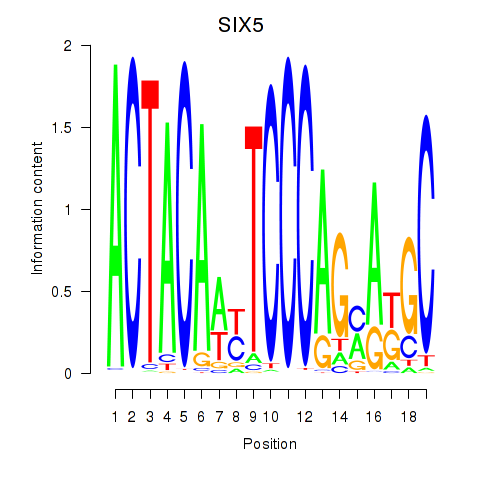
| SIX5_SMARCC2_HCFC1 | -5.653 |
|
|
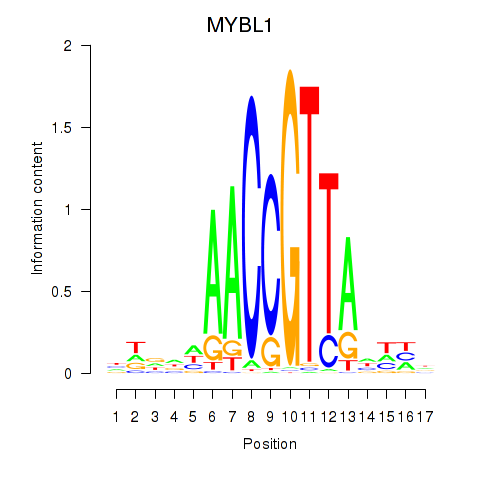
| MYBL1 | -5.738 |
|
|
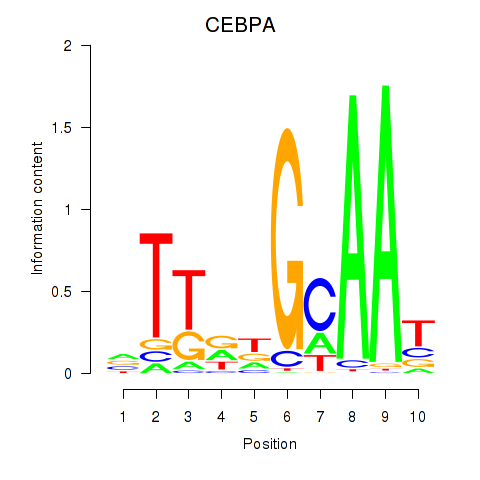
| CEBPA | -6.591 |
|
|
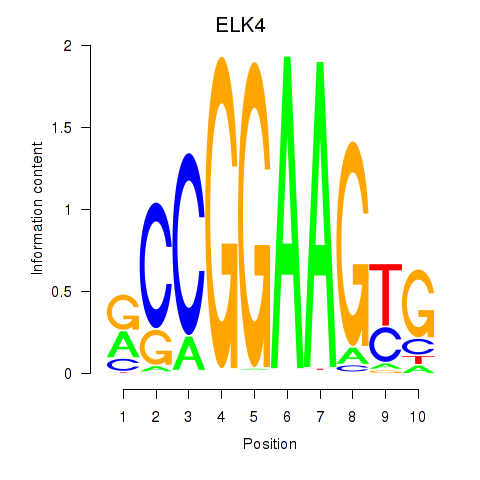
| ELK4_ETV5_ELK1_ELK3_ELF4 | -6.860 |
|
|
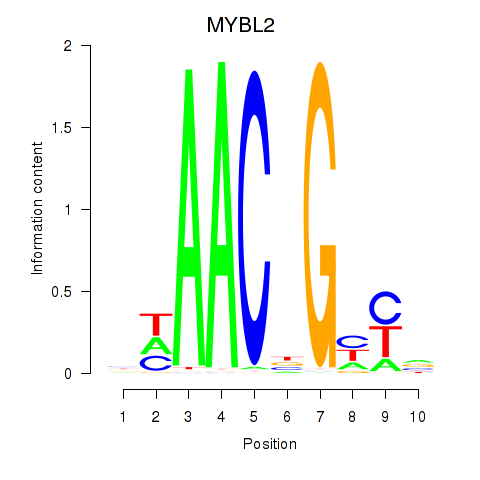
| MYBL2 | -7.605 |
|
|
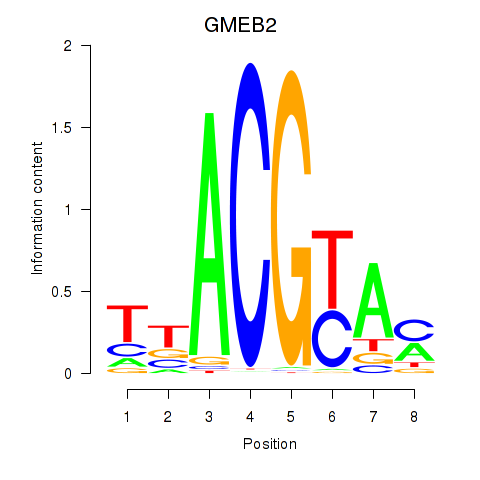
| GMEB2 | -7.743 |
|
|
| SPIC | -8.063 |
|
|
| FOXM1_TBL1XR1 | -8.308 |
|
|
| RUNX1_RUNX2 | -10.013 |
|
|
| GATA3 | -10.528 |
|
|